Thank you for attending Frontiers in Membrane Protein Structural Dynamics 2014

On May 7th, 8th, and 9th, the Membrane Protein Structural Dynamics Consortium (MPSDC) held its second Frontiers in Membrane Protein Structural Dynamics meeting at the Chicago Hilton Hotel. The meeting featured both Consortium members and external invitees, and consisted of eight scientific sessions, poster presentations (and mandatory one-minute Flash! Poster talks), as well as two keynote lectures by Robert Stroud (UCSF) and Klaus Schulten (UIUC). Prior to the conference, the MPSDC hosted several satellite events including a computational modeling workshop, a mini-symposium meeting concerning the latest advances in computational approaches, and a workshop on spectroscopy methodologies. 
By all accounts, the meeting was a tremendous success and participants spoke highly about the scientific discussions that took place. We have made available a brief narrative about the meeting, as well as photos and audio interviews. We asked participants about their research, and their views on Frontiers in Membrane Protein Structural Dynamics 2014, what they felt were some of the highlights of the meeting, and if applicable, their recent collaborations with the MPSDC.
 Access Frontiers... photos and audio interviews » Access Frontiers... photos and audio interviews »
Back to top 
Interview with keynote speaker Klaus Schulten
 Three weeks in advance of Frontiers in Membrane Protein Structural Dynamics 2014, Professor Klaus Schulten (Director of the Computational and Theoretical Biophysics Group at UIUC) phoned in with us from his native Germany to talk about his research interests, the state of the field, the Membrane Protein Structural Dynamics Consortium (MPSDC), and the keynote lecture that he will be giving at the conference. Three weeks in advance of Frontiers in Membrane Protein Structural Dynamics 2014, Professor Klaus Schulten (Director of the Computational and Theoretical Biophysics Group at UIUC) phoned in with us from his native Germany to talk about his research interests, the state of the field, the Membrane Protein Structural Dynamics Consortium (MPSDC), and the keynote lecture that he will be giving at the conference.
In the interview, Dr. Schulten addresses his scientific research interests both past and present, his perspective on some of the key challenges for the field of membrane protein biophysics in the coming 5-10 years, his keynote lecture, and the Membrane Protein Structural Dynamics Consortium.
In addition, Dr. Schulten has made available the movie titled "Photosynthetic Membrane of Purple Bacteria - A Clockwork of Proteins and Processes" which he showed during his keynote lecture, now complete with audio narration taken from the lecture. View the movie:
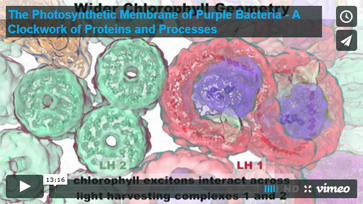
 Read the interview » Read the interview »
Back to top 
Anatrace Poster & Travel Award recipients announced


As previously announced, Anatrace has graciously sponsored travel awards and poster prizes for students and postdocs attending the meeting. Four travel awards ($500) and two poster prizes were decided by a panel on the basis of merit. Winners of these awards were announced at the meeting, and can now be released to the public.
The winners of the Anatrace poster & travel awards are:
Poster Awards:
Anne Georges, Washington University of St. Louis
Investigating the inhibitory effects of a novel monobody on EmrE transport activity
Zachary James, University of Minnesota
EPR Detects Changes in the Transmembrane Region of the SERCA-PLB Complex Upon Ser16 Phosphorylation
Travel Awards:
Adam Chamberlin, University of Calgary
The gating pathway in the voltage-gated proton channel
Michael V. LeVine, Weill Cornell Medical College
NbIT - a new information theory-based analysis of allosteric mechanisms reveals residues that underlie function in the leucine transporter
Nicholas Woodall, University of California, Los Angeles
The positive-inside rule is a local effect
Emilia Ling Wu, University of Kansas
E. coli Outer Membrane and Interactions with OmpLA
Congratulations to all recipients!
Back to top

Satellite workshops and mini-symposium highlight the latest advances in computational modeling and spectroscopy
 As in previous years, the MPSDC sponsored several satellite events in Chicago area prior to Frontiers in Membrane Protein Structural Dynamics 2014. This year, the Computational Modeling Core hosted a membrane protein modeling workshop and a mini-symposium concerning the latest advances in computational approaches to the study of membrane proteins. The modeling workshop provided attendants with an overview of the use of the modeling dynamics and visualization software NAMD and VMD, the CHARMM-GUI Ligand Binder module, and force field parameterization tools. Workshop speakers included Emad Tajkhorshid (UIUC), Wonpil Im (University of Kansas), and Jeff Klauda (University of Maryland). The lecture notes for this workshop are available on our website. This year’s Computational mini-symposium covered a number of topics including methodologies and their applications, voltage gating, and pumps and transporters. As in previous years, the MPSDC sponsored several satellite events in Chicago area prior to Frontiers in Membrane Protein Structural Dynamics 2014. This year, the Computational Modeling Core hosted a membrane protein modeling workshop and a mini-symposium concerning the latest advances in computational approaches to the study of membrane proteins. The modeling workshop provided attendants with an overview of the use of the modeling dynamics and visualization software NAMD and VMD, the CHARMM-GUI Ligand Binder module, and force field parameterization tools. Workshop speakers included Emad Tajkhorshid (UIUC), Wonpil Im (University of Kansas), and Jeff Klauda (University of Maryland). The lecture notes for this workshop are available on our website. This year’s Computational mini-symposium covered a number of topics including methodologies and their applications, voltage gating, and pumps and transporters.
 This year, the Consortium also held its first workshop on Spectroscopy. This workshop was oriented towards young investigators who wish to learn techniques needed for real time monitoring. Talks addressed techniques in solid-state and solution NMR, EPR including DEER approaches, infrared, fluorescence and single molecule techniques including magnetic tweezers. Speakers included Yeon-Kyun Shin (Iowa State), Ana Correa (University of Chicago), Tae-Young Yoon (KAIST), Martin Zanni (University of Wisconsin), Marc Baldus (Utrecht University), Gary Lorigan (Miami University), and Danielle Goldfarb (Weizmann Institute). This year, the Consortium also held its first workshop on Spectroscopy. This workshop was oriented towards young investigators who wish to learn techniques needed for real time monitoring. Talks addressed techniques in solid-state and solution NMR, EPR including DEER approaches, infrared, fluorescence and single molecule techniques including magnetic tweezers. Speakers included Yeon-Kyun Shin (Iowa State), Ana Correa (University of Chicago), Tae-Young Yoon (KAIST), Martin Zanni (University of Wisconsin), Marc Baldus (Utrecht University), Gary Lorigan (Miami University), and Danielle Goldfarb (Weizmann Institute).
Both workshops and minisymposium were well attended and productive, and as before, we will continue to host such satellite events in the future.
Back to top

Two new webservers made available by Computational Modeling Core
The MPSDC Computational Modeling Core has recently supported the development of two new webservers, which are made available for usage by the broader scientific community.
ANMPathway: Find transition pathway between two protein conformations
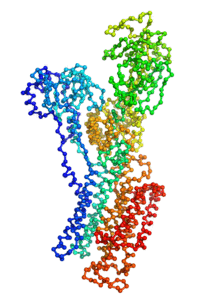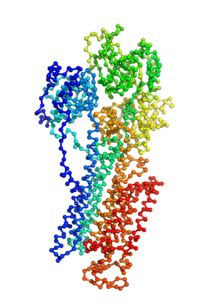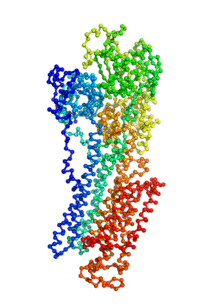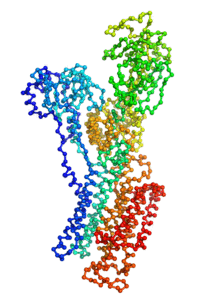 Biomolecular conformational transitions are essential to biological functions. Most experimental methods report on the long-lived functional states of biomolecules, but information about the transition pathways between these stable states is generally scarce. Such transitions involve short-lived conformational states that are difficult to detect experimentally. For this reason, computational methods are needed to produce plausible hypothetical transition pathways that can then be probed experimentally. Here we propose a simple and computationally efficient method, called ANMPathway, for constructing a physically reasonable pathway between two endpoints of a conformational transition. We adopt a coarse-grained representation of the protein and construct a two-state potential by combining two elastic network models (ENMs) representative of the experimental structures resolved for the endpoints. Biomolecular conformational transitions are essential to biological functions. Most experimental methods report on the long-lived functional states of biomolecules, but information about the transition pathways between these stable states is generally scarce. Such transitions involve short-lived conformational states that are difficult to detect experimentally. For this reason, computational methods are needed to produce plausible hypothetical transition pathways that can then be probed experimentally. Here we propose a simple and computationally efficient method, called ANMPathway, for constructing a physically reasonable pathway between two endpoints of a conformational transition. We adopt a coarse-grained representation of the protein and construct a two-state potential by combining two elastic network models (ENMs) representative of the experimental structures resolved for the endpoints.
ANMPathway determines the most probable (lowest energy) transition pathway between two stable endpoints of a conformational transition. Conceptually, ANMPathway represents a direct application of the string method to a two-state CG system approximated by ANM energy surfaces. The result is a sequence of PDB structure (a “string”) smoothly linking the two endpoints.
This webserver was developed by Avisek Das and Sunhwan Jo from Benoit Roux’s group at the University of Chicago, with support from the MPSDC.
 Visit the ANMPathway webserver » Visit the ANMPathway webserver »
LBT-Pred: Insert lanthanide-binding peptide tags into your protein
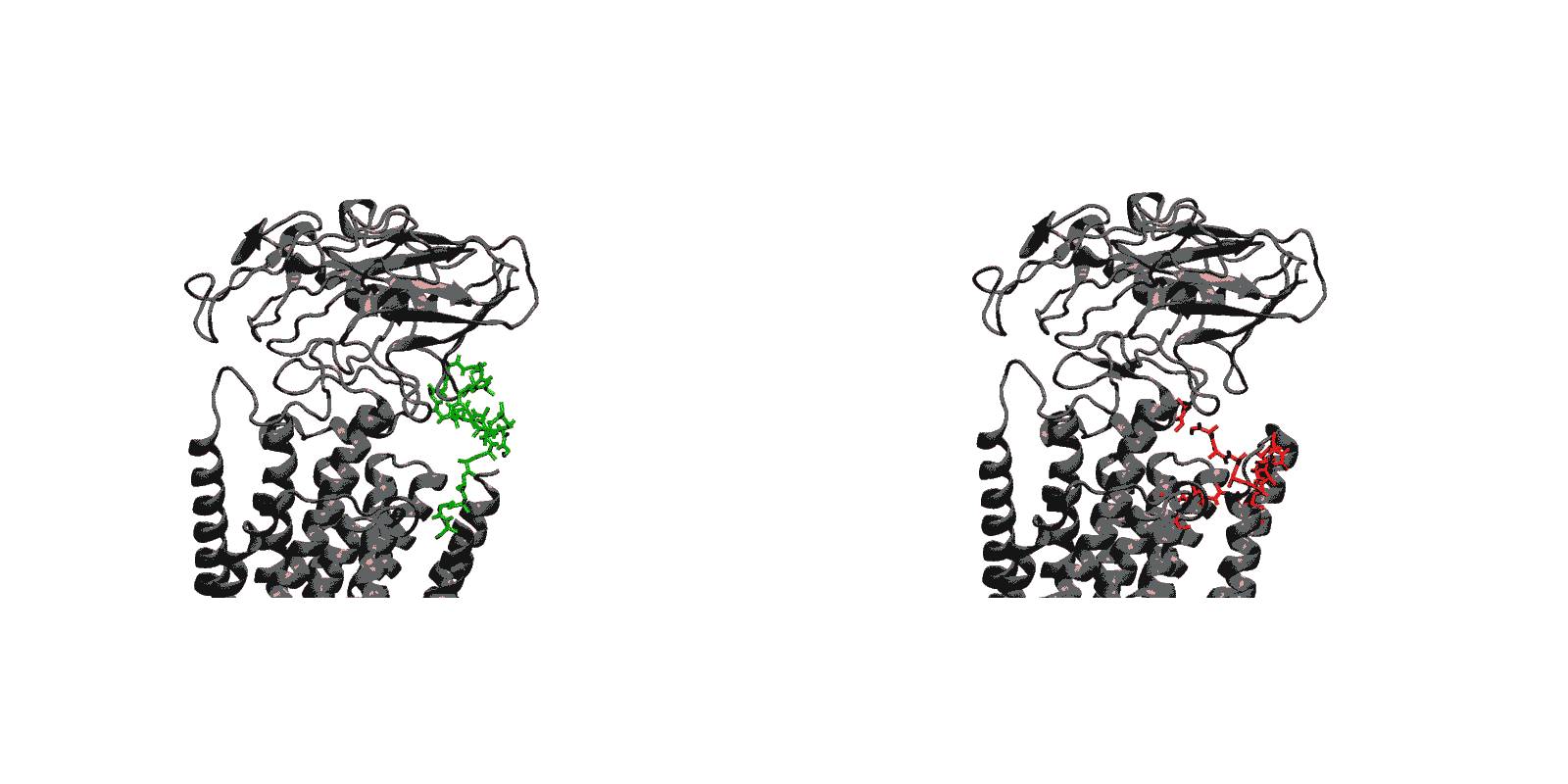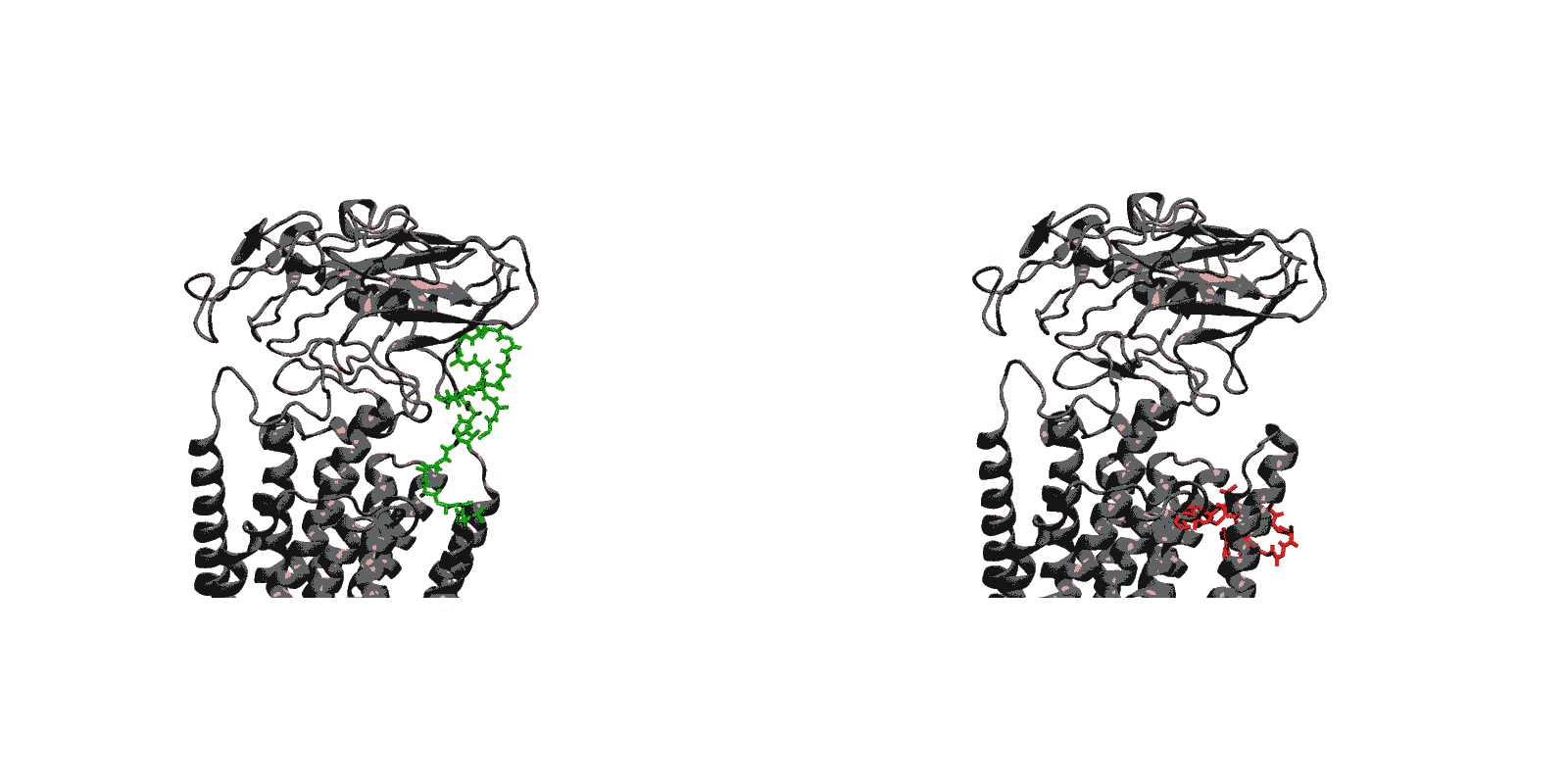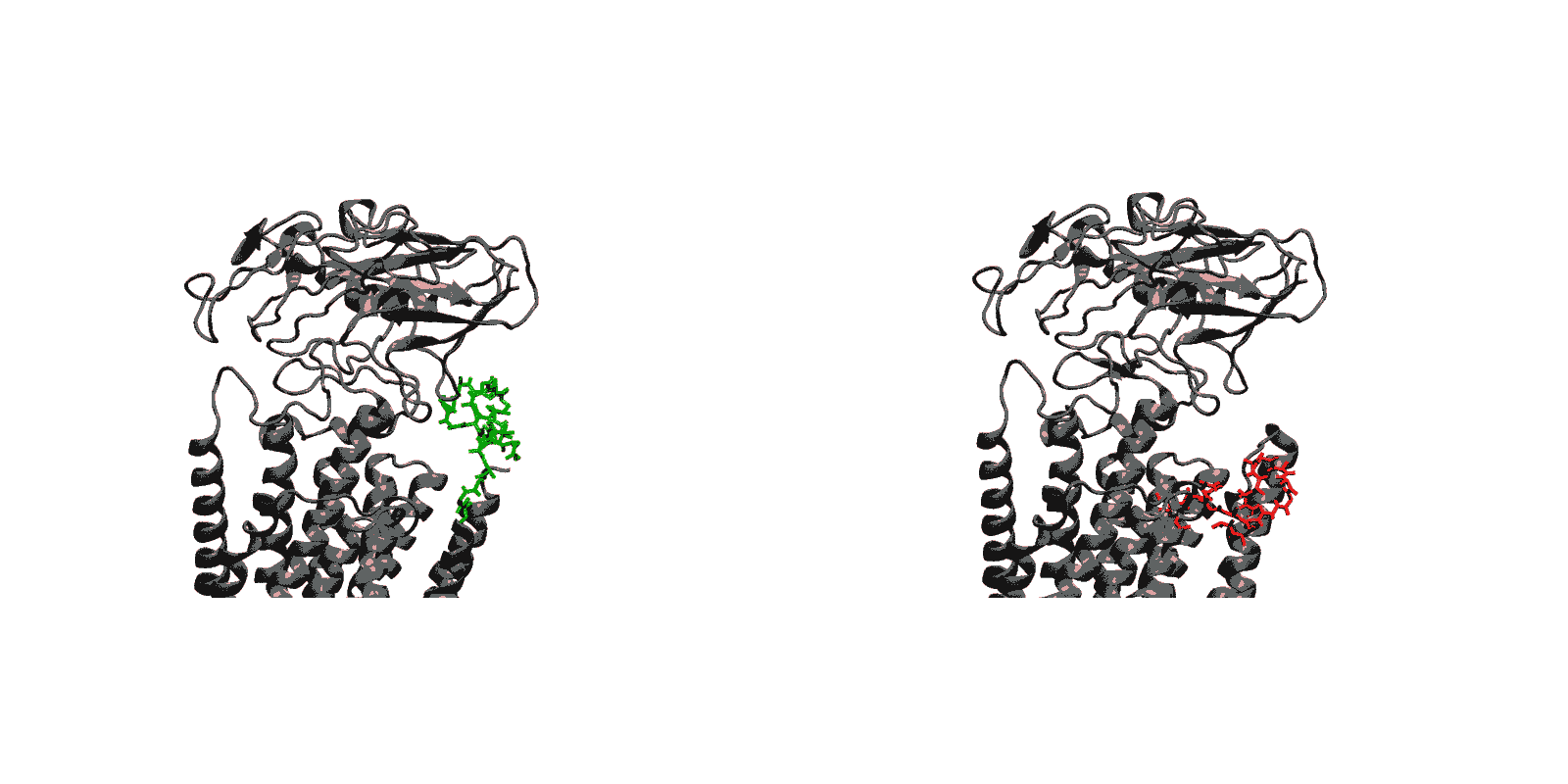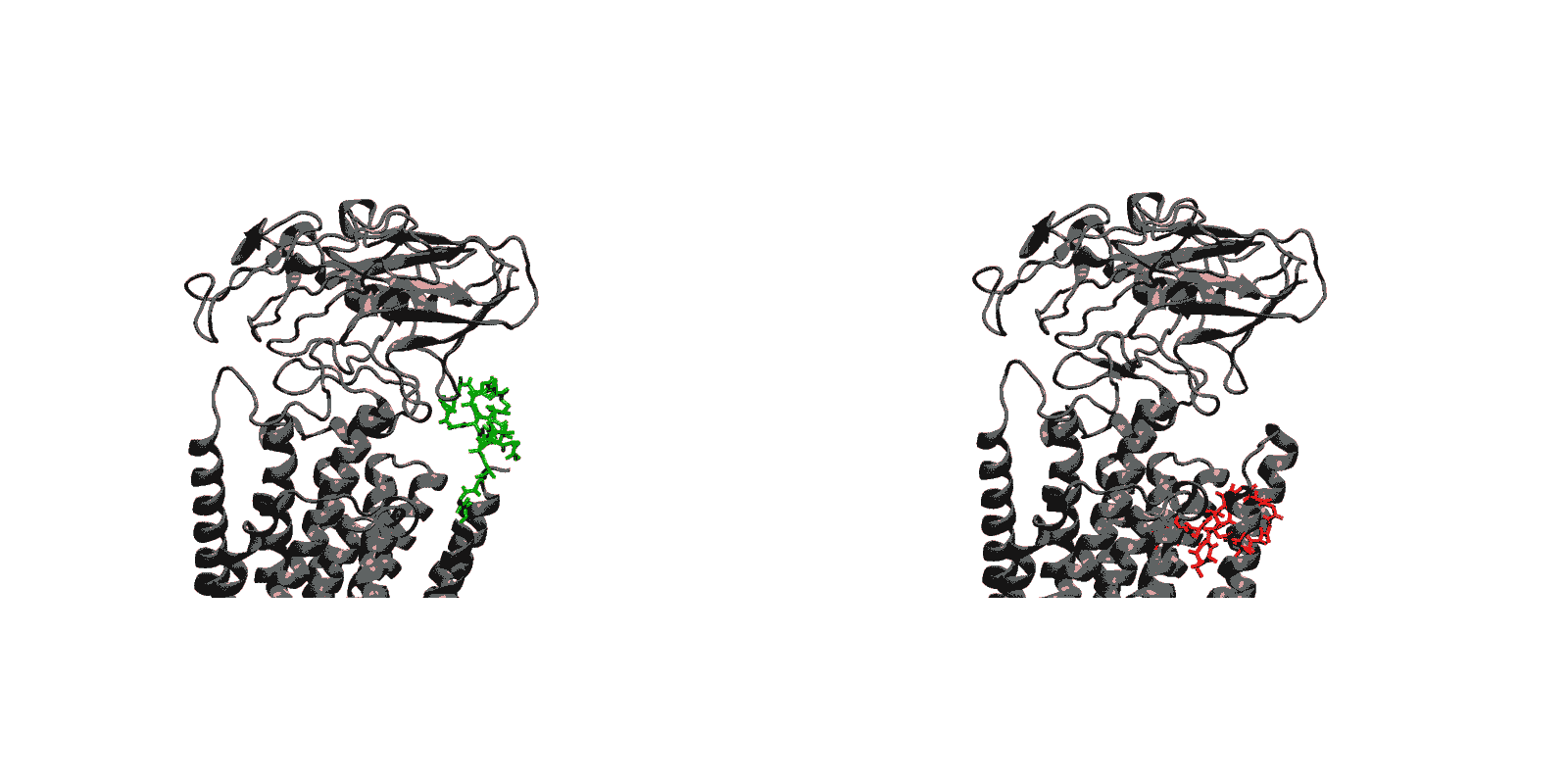
In recent years, high-throughput genomic and proteomic projects have been revealing new genetic information at a very fast pace. The proper interpretation of such information in the context of the underlying biological mechanisms generally requires studies on a molecular level through biophysical and biochemical experiments as well as biomolecular modeling. Hence, the use of biophysical probes that can be easily integrated into protein structures for functional and structural characterization is becoming increasingly common. Lanthanide-binding tags (LBTs) are a class of such probes that are optimized for binding lanthanide ions.
We have created a fully automated webserver for the prediction of LBT insertions onto existing protein structures. The underlying method uses a coarse-grained backbone+Cβ representation of the protein and samples on the backbone dihedral angles. The method predicts the proper conformation of the LBT tag with respect to the parent protein in existing fusion crystal structures. On multiple membrane protein systems, the method’s prediction of feasibility of LBT insertion on various sites qualitatively agree with the experimental data. The method currently reports several metrics to assess the quality of LBT insertion in a specific site in a parent protein. We expect the method to serve as a useful computational tool for experimentalists by helping them select reasonable sites in a given parent protein where the LBT insertion will likely be successful.
This webserver was developed by Aashish Adhikari from Tobin Sosnick’s group at the University of Chicago, with support from the MPSDC.
 Visit the LBT-Pred webserver » Visit the LBT-Pred webserver »
Back to top

ABC Transporter paper by Tajkhorshid laboratory recommended on F1000Prime
One of our long-term projects on Structural Dynamics of ABC Transporters integrates computational, biochemical and spectroscopic approaches to understand the structural dynamics of the ATP binding cassette (ABC) transporters, which are associated with a number of human pathologies and play critical roles in the removal of cytotoxic agents.
Among the MPSDC participants in this project is Dr. Emad Tajkhorshid, whose contribution is applying molecular dynamics (MD) simulations integrating experimental constraints to develop structural models for key conformational states and characterize their inter-conversion during the transport cycle.
Tajkhorshid, together with postdoctoral researcher Mahmoud Moradi, recently published a paper on conformational transitions of ATP exporters which was recommended on the F1000Prime or "Faculty of 1000"
website. F1000 is a team of 5,000 Faculty Members – senior scientists and leading experts in all areas of biology and medicine — plus their associates who provide recommendations of important scientific articles, rating them and providing short explanations for their selections.
 Mahmoud Moradi and Emad Tajkhorshid. Photo taken from the University of Illinois News Bureau website. Photo by L. Brian Stauffer.
Mahmoud Moradi and Emad Tajkhorshid. Photo taken from the University of Illinois News Bureau website. Photo by L. Brian Stauffer.
The publication by Moradi and Tajkhorshid, titled “Mechanistic picture for conformational transition of a membrane transporter at atomic resolution“, was published on November 19, 2013 in PNAS vol. 110, no. 47. The paper describes a nonequilibrium approach which they developed to characterize the conformational transition of MsbA, a member of the ATP-binding cassette exporter family, which is involved in transport of diverse substrates across the membrane. The F1000Prime recommendation, written by Qian Cui, tagged the publication as Good for Teaching, as having an Interesting Hypothesis, and for Technical Advance. Read the F1000 recommendation »
The article was also featured on the popular NIH Biomedical Beat blog, and the University of Illinois News Bureau. Additionally, produced a video in which Tajkhorshid describes how his laboratory has successfully simulated the molecular dance moves that a multidrug resistance membrane transporter undertakes as it pumps compounds out of a cell. Watch the video:
 Read more » Read more »
Back to top

CLC Channel/Transporter Family Project team combines computational and experimental methodologies in PNAS publication
The Conformational Dynamics in the CLC Channel/Transporter Family Project of the Membrane Protein Structural Dynamics Consortium (MPSDC) has published its first publication titled “Water access points and hydration pathways in CLC H+/Cl- transporters” in Proceedings of the National Academy of Sciences of the United States of America (PNAS). This Consortium Project is spearheaded by Principal Investigator Merritt Maduke. The laboratories of Marc Baldus, Emad Tajkhorshid, and Hassane Mchaourab are also collaborators in the Project’s ongoing research.

CLC transporters are biologically essential proteins that catalyze the transmembrane exchange of chloride for protons. The permeation pathway for chloride through the transporters has been well characterized. In this publication, Han et al. study the more elusive permeation pathway for protons. Through computational modeling, they show that water molecules can permeate deep inside the protein and form continuous wires. To test the hypothesis that these water wires mediate proton transport, they mutated residues predicted to impede water wire formation and experimentally evaluated the effects of the mutations. The results from their concerted computational and experimental approach strongly support the role of water in proton transport by CLCs and provide a valuable framework for investigating their overall transport mechanism.
In a commentary piece published by PNAS this month, Mounir Tarek (National Center of Scientific Research at the University of Lorraine, France) describes the significance of this paper for future research of chloride channels, and highlights the fruitful combination of simulation and experimentation: “In PNAS, Han et al. used molecular dynamics (MD) simulations of the CLC-ecl, a CLC exchanger from Escherichia coli to specifically address this issue. The predictions of their calculations were tested by additional experiments, providing a robust description of the molecular prerequisites to proton transport in CLC-ecl and a framework for refining models of the Cl-/H+-coupled transport in CLCs.”
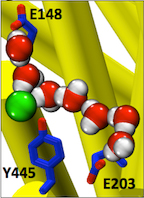 Indeed, this research article reports results from the tightly coordinated efforts of a computational and an experimental lab brought together by the Consortium. The study addresses a critical question about the CLC transporter mechanism: how does H+ traverse the hydrophobic expanse of the CLC protein? The MD simulations performed reveal water dynamics, water-wire formation, and side-chain conformational change not observed in any of the static crystal structures. The functional analyses validated predictions of the simulations and confirm the importance of water dynamics in the transport mechanism. The simulations further reveal that Cl- binding is critical for water-wire formation, thus providing a satisfying explanation for the puzzling experimental observation that Cl- occupancy correlates with the ability of CLCs to transport H+. These studies provide a crucial framework for understanding how H+ and Cl- binding and translocation steps are coordinated in the CLC transporters to control stoichiometric transport. Indeed, this research article reports results from the tightly coordinated efforts of a computational and an experimental lab brought together by the Consortium. The study addresses a critical question about the CLC transporter mechanism: how does H+ traverse the hydrophobic expanse of the CLC protein? The MD simulations performed reveal water dynamics, water-wire formation, and side-chain conformational change not observed in any of the static crystal structures. The functional analyses validated predictions of the simulations and confirm the importance of water dynamics in the transport mechanism. The simulations further reveal that Cl- binding is critical for water-wire formation, thus providing a satisfying explanation for the puzzling experimental observation that Cl- occupancy correlates with the ability of CLCs to transport H+. These studies provide a crucial framework for understanding how H+ and Cl- binding and translocation steps are coordinated in the CLC transporters to control stoichiometric transport.
 Read more » Read more »
Back to top
|
