Accelerating Membrane Insertion of Peripheral Proteins with a Novel Membrane Mimetic Model
By Y. Zenmei Ohkubo, Taras V. Pogorelov, Mark J. Arcario, Geoff A. Christensen and Emad Tajkhorshid
Published in Biophysical Journal 102(9): 2130-2139 on May 2, 2012. PMID: 22824277. PMCID: PMC3341550. Link to Pubmed page.
Core Facility: Computational Modeling
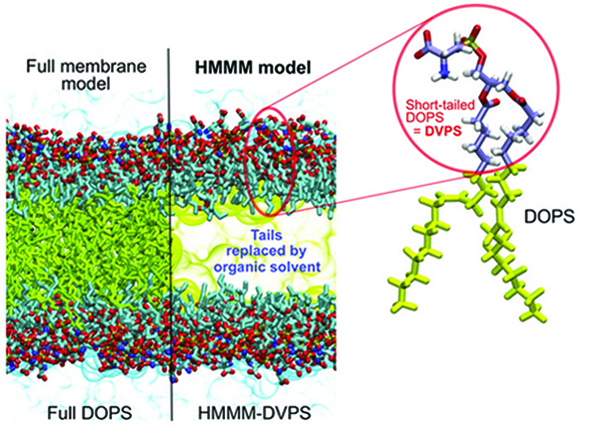
Figure 1. Contrasting the HMMM and conventional representations of a membrane. In an HMMM model, a large fraction of the acyl tails (left, yellow sticks) of the membrane-forming lipids is replaced by a liquid organic phase (right, yellow area). In this study, a full DOPS lipid molecule (inset) is represented by a short-tailed DVPS molecule (the circled fraction of the molecule), and the space vacated by the removal of the C6–C18 portion of the acyl tails is filled with an organic solvent (yellow). Oxygen atoms are red, nitrogen blue, phosphorus gold, and carbon ice blue, except the C6–C18, which are in yellow. Bulk water molecules are shown in light blue.
Abstract
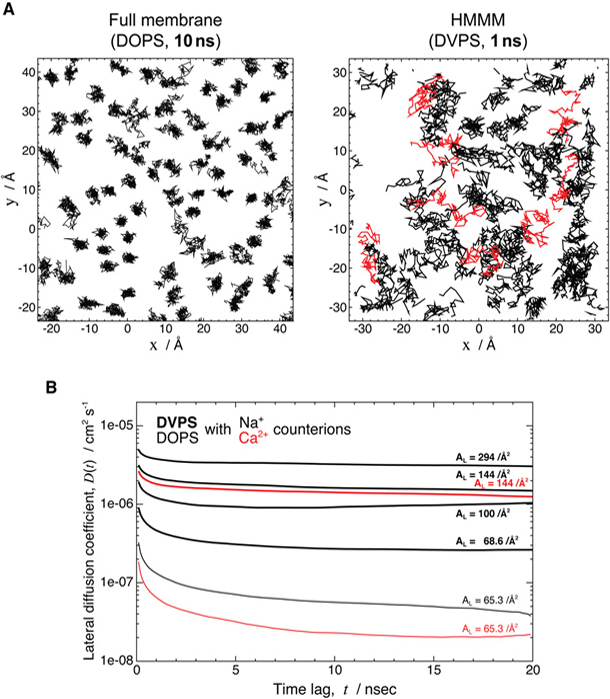
Characterizing atomic details of membrane binding of peripheral membrane proteins by molecular dynamics (MD) has been significantly hindered by the slow dynamics of membrane reorganization associated with the phenomena. To expedite lateral diffusion of lipid molecules without sacrificing the atomic details of such interactions, we have developed a novel membrane representation, to our knowledge, termed the highly mobile membrane-mimetic (HMMM) model to study binding and insertion of various molecular species into the membrane. The HMMM model takes advantage of an organic solvent layer to represent the hydrophobic core of the membrane and short-tailed phospholipids for the headgroup region. We demonstrate that using these components, bilayer structures are formed spontaneously and rapidly, regardless of the initial position and orientation of the lipids. In the HMMM membrane, lipid molecules exhibit one to two orders of magnitude enhancement in lateral diffusion. At the same time, the membrane atomic density profile of the headgroup region produced by the HMMM model is essentially identical to those obtained for full-membrane models, indicating the faithful representation of the membrane surface by the model. We demonstrate the efficiency of the model in capturing spontaneous binding and insertion of peripheral proteins by using the membrane anchor (γ-carboxyglutamic-acid-rich domain; GLA domain) of human coagulation factor VII as a test model. Achieving full insertion of the GLA domain consistently in 10 independent unbiased simulations within short simulation times clearly indicates the robustness of the HMMM model in capturing membrane association of peripheral proteins very efficiently and reproducibly. The HMMM model will provide significant improvements to the current all-atom models by accelerating lipid dynamics to examine protein-membrane interactions more efficiently.