The Transport Cycle in Neurotransmitter Uptake Systems
Core Facilities: Computational Modeling – Synthetic Antigen Binder (SAB) Generation and Crystallography – Membrane Protein Expression/Purification
Using the recently determined crystal structure of a prokaryotic leucine transporter (LeuT), this Project will explore the conformational changes and dynamic properties relevant to function in Neurotransmitter transporters translocation cycle using a combination of computational, functional, and spectroscopic approaches.
Project Description
Using the recently determined crystal structure of a prokaryotic leucine transporter (LeuT), this Project will explore the conformational changes and dynamic properties relevant to function in Neurotransmitter transporters translocation cycle using a combination of computational, functional, and spectroscopic approaches.
Eukaryotic neurotransmitter:Na+ symporters (NSS) terminate neurotransmission through uptake of various neurotransmitters, including biogenic amines (dopamine, norepinephrine, and serotonin) and amino acids (GABA and glycine). The transporters for dopamine (DAT), norepinephrine (NET), and serotonin (SERT), are the primary targets for psychostimulant drugs such as cocaine and amphetamine, and for many antidepressants. Genes encoding more than 200 putative NSS homologs have been computationally identified in prokaryotes genomes, thereby expanding the functional spectrum of this transporter family. The elucidation of the high resolution structure of the
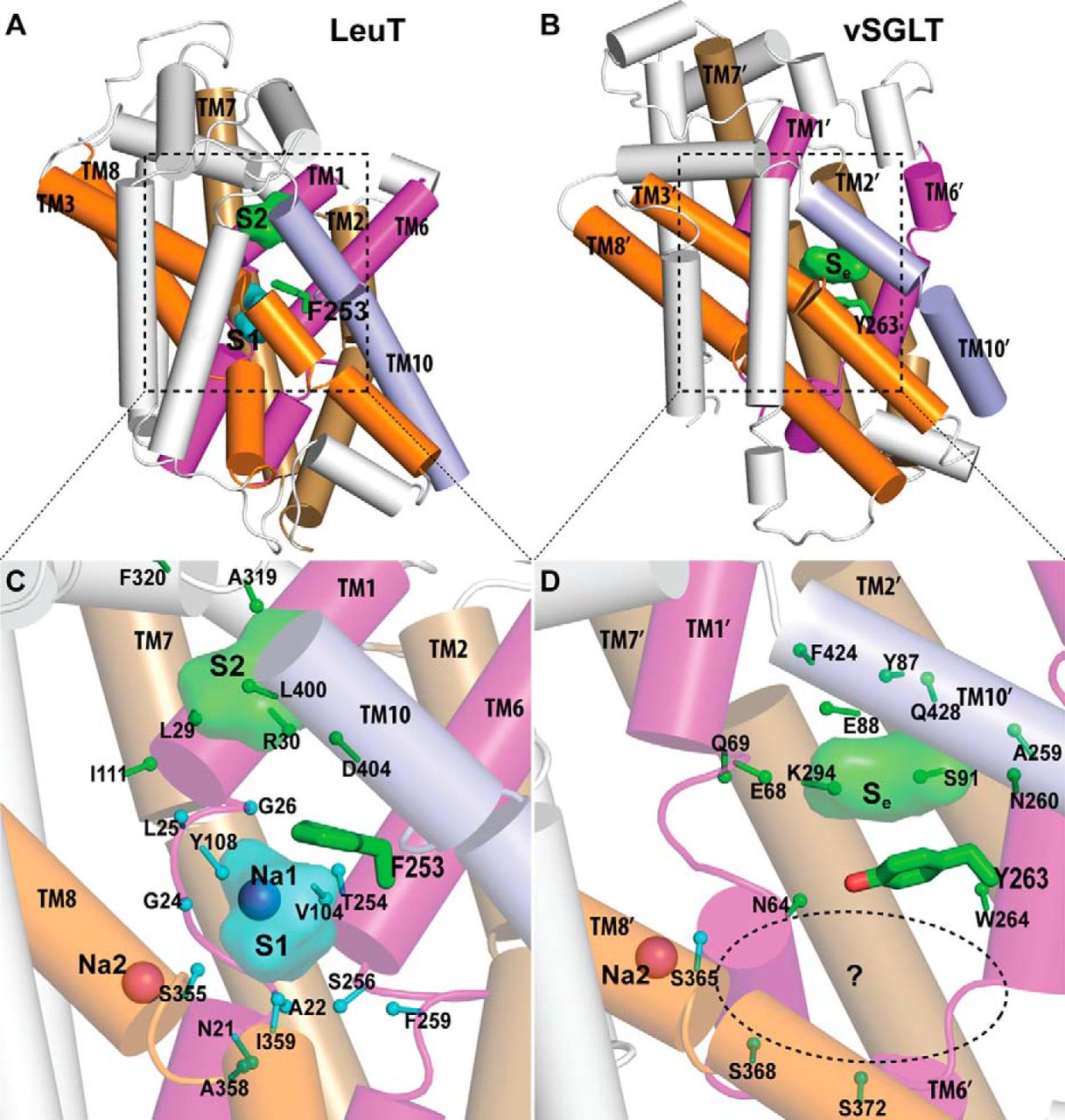
NSS homolog LeuT, an amino acid transporter from Aquifex aeolicus, was a major advance towards understanding structure-function relationships of NSS-proteins. The LeuT crystal structure revealed an occluded state in which one substrate, leucine, and two Na+ are bound, but provided limited clues to the molecular mechanism of transport. In particular, the key functional feature of secondary transport – how the energy of ion gradients (in this case Na+) is converted into the uphill accumulation of substrate – cannot be deduced directly from existing crystal structures, which now include a number of inhibitor bound forms of LeuT, andother transporters with LeuT-like folds (vSGLT,Mhp1, BetP, and AdiC).
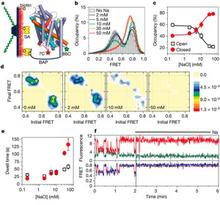
The characterization of the conformational rearrangements that must be associated with transport by NSS was aided by steered molecular dynamics simulations that explored the extracellular substrate translocation pathway of LeuT. A remarkable finding is the identity and key mechanistic role of a second substrate (S2) binding site in the extracellular vestibule that is comprised of residues shown to participate in binding tricyclic antidepressants (TCAs). Because the role of this site has not been well understood, we carried out an extensive series of experiments and computations that revealed that the two binding sites can be occupied simultaneously by substrate; substrate binding in the S2 site allosterically triggers intracellular release of Na+ and substrate from the primary (S1) site. In this mechanism, that is consistent with the results from simulations, substrate bound in the S2 site functions as a “symport effector”. This distinguishes the substrate from TCAs, which bind in the same vestibule but do not promote substrate release from the S1 site. In fact, TCAs competitively block substrate binding to the S2 site, thereby inhibiting transport as “symport uncouplers.”
Although binding of substrate and TCAs to the S2 site must lead to quite different large-scale conformational changes, the LeuT crystal structures with and without TCA are very similar. The important consideration in the interpretation of this situation is that the second substrate, now known from our studies to be of key importance as an allosteric factor in function, was not present in the original crystal structure due to the conditions employed. Efforts by us and others to crystallize LeuT with a substrate-occupied S2 site have failed. However, our recent crystallographic and functional approaches clearly identified a molecule of octylglucoside, the detergent used for all crystallization of LeuT to date, in the S2 site of LeuT where it prevents substrate binding and acts as an inhibitor, just like bound TCAs and SSRIs. We thus conclude that all present LeuT crystalother approaches to characterize the functional dynamics of these proteins. To this end, we undertook collaborative studies using electron paramagnetic resonance (EPR) spectroscopy of site-directed spin-labeled (SDSL) LeuT, in parallel with, and in order to probe the result of the computational simulations of the various states. From the initial findings of this work we have already observed evidence of (i) altered conformational states in the presence of Na+ and substrate; and (ii) changes in distance between pairs of spin labels in different states. We hypothesize that a symport effector triggers conformational changes propagated from the extracellular to the intracellular side that involve hydration and dehydration of the sites, whereas a symport uncoupler blocks these events, thereby preventing transport. Based on this hypothesis we propose to identify the conformational changes and dynamic properties relevant to function in the transport cycle using the powerful and mutually complementary combination of computational, functional, and EPR approaches we have established.