By Eduardo Perozo and Hassane Mchaourab
The functional behavior of most proteins is intimately related to their conformational dynamics. It follows that a thorough understanding of structure-function relations in a given protein requires a molecular description of the types and extent of the protein movements underlying function. This includes catalysis, transport, cellular locomotion, regulation of activity, and formation of protein assemblies. X-ray and NMR structures have made it clear that proteins undergo a wide variety of motions, whether as a consequence of a triggered action (ligand binding, protein-protein contact, physical stimuli) or as the result of the natural fluctuations around a flexible linker between domains. Closer examination reveals that the types and dynamic behavior of these movements can be hierarchically divided according to four major groups (Figure 1): Side-chain rotamers (ps-ns), reorientation of secondary structure elements (ns), large domain movements (ns-ms), and whole subunit rearrangements in cooperative oligomeric complexes (ms-ms).
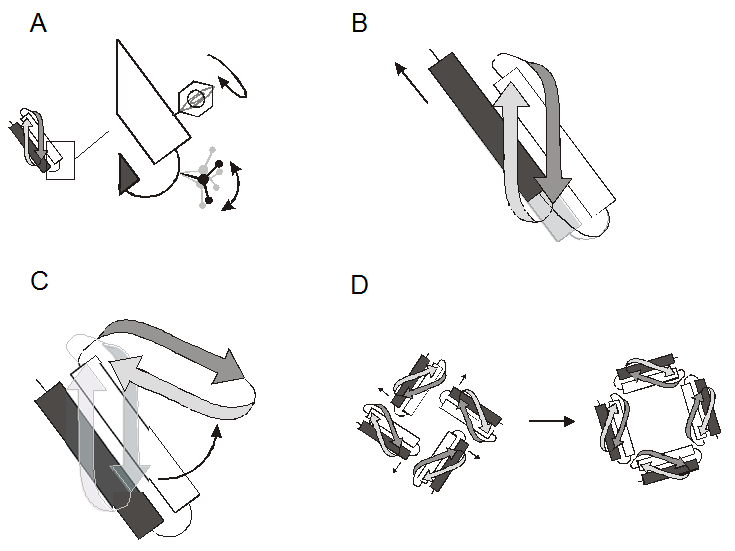
Figure 1 (click to enlarge)
The types of molecular motions in proteins. A. Side-chain rotamers, B. Reorientation of secondary structure elements, C. Large domain motions, D. Whole subunit rearrangements.
Among the types of mechanisms of motion mentioned above, by far, the best understood is that occurring among protein domains. Gerstein and co-workers have reviewed and classified domain motions from a growing database of proteins with known three-dimensional structures (Gerstein et al., 1994). They concluded that most domain motions ultimately originate from two basic mechanisms, hinge bending or shear motions. In hinge bending, motion occurs due to changes in a few main-chain torsion angles along a localized region of the protein connecting two interacting modules (strands, b-sheets, a-helices, or combinations of them). These two modules are normally not constrained by tertiary packing interactions, and the transition from “open” to “closed” positions is accompanied by the exclusion of water molecules from the inter-domain space. In contrast, interactions among tertiary structure elements are severely constrained due to packing interactions, precluding large changes in main-chain torsion angles. Hence, shear motions have been observed mostly among closely packed regions of proteins. Shear involves small changes in side-chain torsion angles among interdigitating side-chains, without large changes in main chain conformation. These can be observed either parallel or perpendicular to the interface of closely packed segments of a polypeptide chain. An argument is also made that because these types of protein rearrangements are usually fast, transition between the two conformations cannot involve large energy barriers. This is particularly relevant in the analysis of equilibrium fluctuations, since the relatively weak forces involved in crystal packing are known to favor one of the two possible conformations, as in the case of the T4 lysozyme (see below). A number of useful databases of protein motions continue to be maintained on the web by Gerstein and co-workers and by Hayward and coworkers.
It can be argued that one of the most powerful aspects of site-directed spin labeling as a structural approach, is its potential to discern the types of protein motions described above. Information on the relative movement of secondary structure elements, domains or whole subunits can be obtained by studying patterns of change in probe mobility and solvent accessibility or changes in relative distance among residue pairs between domains or subunits. However, because this is a reporter group technique, side-chain rotamer conformations cannot be analyzed.
Animation courtesy of Dr. Peter Fajer, Florida State University.
Analysis of equilibrium fluctuations
Protein conformational equilibria are the results of the thermally activated interconversion of the structure between multiple states of similar energies. These motions differ in their amplitude, time scale and biological consequence. Thermally activated equilibrium fluctuations in the structure can facilitate access of a substrate to an active site cleft, binding of an effector to an allosteric site and transport of ions through channels. Because thermally activated equilibria involve states of similar energies, lattice forces may be sufficiently large to select a particular state in a crystal, obscuring the existence of others. In some cases, crystallization of the same protein in two different crystal forms allows the observation of multiple conformations. Dynamics can also be inferred from the occurrence of disorder in the crystal and analysis of thermal factors.
Conformational equilibria in solution can be explored by SDSL as long as the interconversion rate is within the time scale of the employed EPR technique. Since the dynamic range of EPR covers over 7 orders of magnitude, it provides a suitable window for many protein motions of interest. Both the mobility of the nitroxide side chain and the distance between the nitroxide and a second paramagnetic center can be used to detect conformational changes. The structural determinants of these observables as well as their resolution makes them ideal to detect large scale concerted motion of large segments of the protein molecule. The population of multiple structures results in a heterogeneous environment in the vicinity of the nitroxide that is reflected in the EPR spectral lineshape as multiple motional states. Similarly, if the dynamics involve rigid body motion of a secondary structural element it will modulate the proximity of an attached nitroxide to a nearby reference paramagnetic center. Although the contribution of the backbone flexibility to the mobility of the nitroxide is not well understood, in principle backbone fluctuations are also detectable. Because conformational equilibria involve motions that are constantly occurring in solutions, the spectral properties are ensemble averages with each state with distinct spectral characteristics contributing to a different degree.
In practice, detailed quantitative interpretation of EPR data arising from such ensembles are yet to be developed. At the fundamental level, multicomponent EPR spectra can arise from different conformations of the spin label itself. Furthermore, the addition of parameters such as the number of intermediate states and the mobility and/or internitroxide distance in each state makes the linear CW-EPR spectrum underdetermined.
The problem is considerably simplified if the structure of one of the conformers is known. This provides a reference state against which spectral observables can be compared. An ideal situation arises if the states of known structure can be selectively populated via changes in the physicochemical conditions or by binding of a ligand or a substrate. In this case, reference EPR spectra can be obtained and the sign of the changes in mobility or interresidue distances when multiple states are populated can be interpreted in terms of the nature, extent and magnitude of the movement involved in the equilibria.












