
University of Illinois professor and MPSDC team member Emad Tajkhorshid, along with co-PIs Chad Rienstra (Chemistry) and James Morrissey (Biochemistry) have been awarded a Director’s Transformative Research Award from the National Institutes of Health for their highly creative approach to the study of cell membrane lipids.
Membrane proteins are abundant in eukaryotic cells and play important roles in a great many biological processes ranging from cell adhesion and recognition to energy production to signaling cascades.
Membrane proteins make up more than half of the targets for currently approved drugs, which underscores their relevance to human disease but less is known about the membrane lipids that interact with proteins and ligands.
It is becoming increasingly clear that lipids are effector molecules that modulate and/or directly carry out essential biological functions at very different rates depending on what types of lipids are present. Some examples include blood clotting, cell recognition (in immunological response especially), ion conduction (important for neuronal function and viral infection), transport of drugs across the membrane, and pain response.
A potential long-term application is the development of more effective drugs that target biological membranes. Since about 60% of the drugs on the market target membrane-bound proteins, a better understanding of lipid structure and dynamics could greatly improve the efficacy of drug design efforts by modeling the interactions that take place. This would have broader impacts on understanding all the biological functions above and potentially to address the resulting pathologies or diseases. Better blood thinners would help to ameliorate deep vein thrombosis, heart attacks and strokes. Improved modeling of immunological cell recognition and viral life cycles would help to address infectious diseases ranging from influenza to HIV/AIDS. Understanding how drugs are transported would aid in the development of better antibiotics. The project aims to develop a toolkit of methods that would be available to researchers addressing this range of problems and many others.
The High-Risk, High-Reward Research (HRHR) program, supported by the National Institutes of Health (NIH’s Common Fund) awarded twelve transformative research awards funded by the Director’s office. The awards span the broad mission of the NIH and include groundbreaking research.
Read more about the project here: link




 The 2014 International Biophysics Congress, which is taking place in Brisbane, Australia this year, will have the pleasure of hosting none other than MPSDC Director
The 2014 International Biophysics Congress, which is taking place in Brisbane, Australia this year, will have the pleasure of hosting none other than MPSDC Director 
 At the conference,
At the conference, 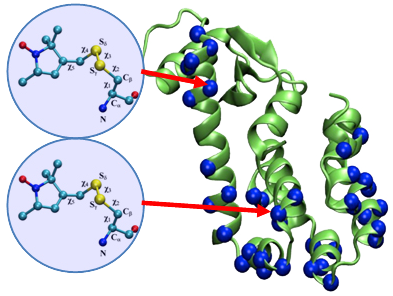 The article co-authored by Benoît Roux and Jeane Weare was highlighted by the Journal of Chemical Physics on their
The article co-authored by Benoît Roux and Jeane Weare was highlighted by the Journal of Chemical Physics on their 






