Structural Dynamics of Hormone-Receptor Action
Core Facilities: Membrane Protein Expression/Purification – Computational Modeling
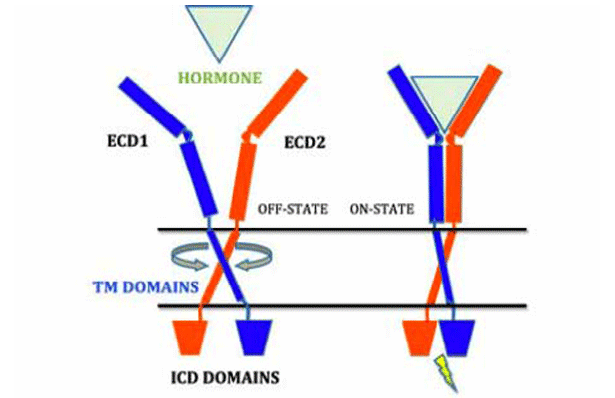
Project Description
Mechanisms and Energetics of Transmembrane-induced Signaling of Cytokine Receptors: A. Kossiakoff (University of Chicago), Wonpil Im (University of Kansas)
How transmembrane (TM) domains of membrane proteins transmit the signal across the cell membrane has long been a subject of interest and challenge in biology. There is a recent paradigm shift in the activation process of the cytokine receptor superfamily. The role of cytokine hormone binding to the extracellular domain is now recognized as an “inducer” of the conformational change of pre-dimerized TM domains that triggers subsequent intracellular responses, which is drastically different from the traditional view as an “organizer” to initiates the receptor TM dimer formation (Fig. 2). Our long-term goal is to delineate the mechanisms and energetics of TM-induced signaling of various single-pass TM receptors during the inactive to active transition upon ligand binding.
Our hypothesis is that the off-state conformation is much more stable than the on-state one. We propose that the major role of ligand binding is mainly to disrupt the pre-dimerized (energetically stable) TM-TM contact that locks-in the off-state structure, to direct the (energetically unstable) active on-state structure. In this project, we will use human growth hormone receptor (hGHR) and human prolactin receptor (hPRLR) in the cytokine receptor superfamily as prototypical model systems for homodimeric TM-induced activation. The objectives are to determine the interfacial residues of hGHR and hPRLR TM dimers and to elucidate the conformational and energetic changes during the activation process by innovative combination of versatile computational and experimental approaches. The successful completion of this project will have a significant impact on the field not only by elucidating the TM signaling mechanism and energetics, but also by providing the computational and experimental methods that can be used to characterize the biological activation process of other cytokine receptors and the plentitude of other single-pass TM receptors, which all have the critical importance to biology and thus human health.
Determine the interfacial residues of hGHR TM dimer. Using replica-exchange molecular dynamics (MD) simulations in the implicit membrane model, we have generated various TM dimer models of hGHR with different helix-helix interfaces and crossing angles. We will test the models by “TOXCAT” experiments with the mutants of putative interfacial residues (Fig. 4). We will also perform all-atom MD simulations of each model in explicit membranes to examine their stability, dynamics, and structural change. The changes in the TM association free energy, measured from TOXCAT experiments, will be further investigated by performing the potential of mean force (PMF) calculations as a function of TM helix distance.
Elucidate the conformational and energetic changes during hGHR activation. hGHR is a particularly appealing system to study TM-induced signaling. This is because the off to on-state switch involves formation of a TM proximal disulfide link that can be accurately simulated using a very simple model system. Therefore, we will perform all-atom MD simulations to characterize the structural changes of each dimer model upon activation by slowly changing the distance of two Cys241 sidechains to bring them to a distance (~4 Å) and geometry to form the disulfide bond. The simulation results will be validated by EPR spectroscopy with site-specific spin labels at the appropriate positions. Then, we will perform the PMF calculations during the off-to-on switch using the helix restraint potentials that we have recently developed.
Determine the mechanism and energetics of hPRLR TM-induced signaling. The hPRLR TM has no sequence homology to its evolutionary related hGHR counterpart. hPRLR has no equivalent cysteine residue to Cys241 of hGHR to form an on-state disulfide link. However, it does contain a Cys residue in its TM sequence. Therefore, we will determine the different structural aspects toward the mechanism and energetics of hPRLR TM-induced signaling based on the proposed approaches in AIM 1 and AIM 2.
This combination of computational and experimental approaches will enable us to address TM-induced signaling of hGHR and hPRLR. Determining the areas of similarity and differences will establish a better theoretical footing for understanding how this large class of receptors operates. The successful completion of this proposed project will have a significant impact on the field not only by elucidating the TM signaling mechanism and energetics, but also by providing the computational and experimental methods that can be used to characterize other cytokine receptors and the plentitude of other single-pass TM receptors.


