Spectroscopy and Instrumentation
Project Description
The main objective of the Membrane Protein Structural Dynamics consortium is to obtain a mechanistic understanding of how the structural dynamics of membrane proteins generate its function. The experimental study of structural dynamics is approached with a variety of spectroscopic methods and biophysical procedures that are in constant development and improvement. The objective of the Instrumentation Spectroscopy Core (SIC) is two-fold: first, to improve presently available instruments, develop new instruments, or methods and procedures aimed at improving on signal to noise ratio, specificity and time resolution of spectroscopic techniques and combining them with functional techniques, such as electrophysiology. The second objective, to provide service to the MPSD members, and the community at large, with the new developments as well as with the spectroscopic and functional techniques presently installed.
Electron paramagnetic resonance (EPR) and fluorescence provide complementary information on the dynamics of structural changes in membrane proteins. As the typical spin labels are small and labeling procedures are relatively simple, established tool kits for steady state and time-domain EPR techniques can provide structural and dynamic information to reveal conformational changes in purified proteins. To follow the time course of conformational changes during function time-resolved EPR techniques, such as rapid freeze quench (RFQ), can be employed. The recent development of a microfluidic-based RFQ apparatus (by Gefen and Goldfarb labs) that uses minimal amounts of sample and high-frequency, pulsed-EPR instrumentation offers the possibility of applying advanced EPR approaches to resolve time-dependent conformational changes in membrane proteins. We will pursue this new and exciting direction in Aim 1.
Fluorescence probes, although larger than EPR probes, can follow structural changes in physiological environments at room temperature and at the single-molecule level. As fluorescence decay is typically in the ns range, it is – in principle – possible to follow dynamics from the ns to second (or longer) time scales. Hence, a key aim of this core is to take advantage of the large range of time scales probed by fluorescence. We will do so by optimizing the probes and perfecting the detection techniques (both ensemble and single-molecule) that enable the tracking of dynamic processes in membrane proteins. While the finite photon flux and photostability of single-fluorophores typically limits single-molecule imaging techniques to the ms regime, we will push this boundary to the µs regime through the development of intramolecularly stabilized organic fluorophores.
Fluorescence emission can be detected on membrane proteins expressed in heterologous systems to reveal structural changes during function. A fluorophore’s excited state lifetime reports on its environment, providing information on the nature of a conformational change. A key aim of this Core is to develop methods to measure fluorophore lifetimes combined with electrophysiology. The Specific Aims of the SIC are:
AIM 1: To further develop and perfect a microfluidic rapid freeze quench (RFQ) EPR system to enable measurements of frozen samples that are generated by rapid mixing of reactants and make RFQ accessible to members of the consortium. This technique will be applied in conjunction with double electron-electron resonance (DEER) and Electron nuclear double resonance (ENDOR) experiments.
AIM 2: To further develop and enhance single-molecule fluorescence techniques:
- Test, expand and make available high-performance organic fluorophores. Test those that are developed with unnatural amino acid technologies in the protein production core.
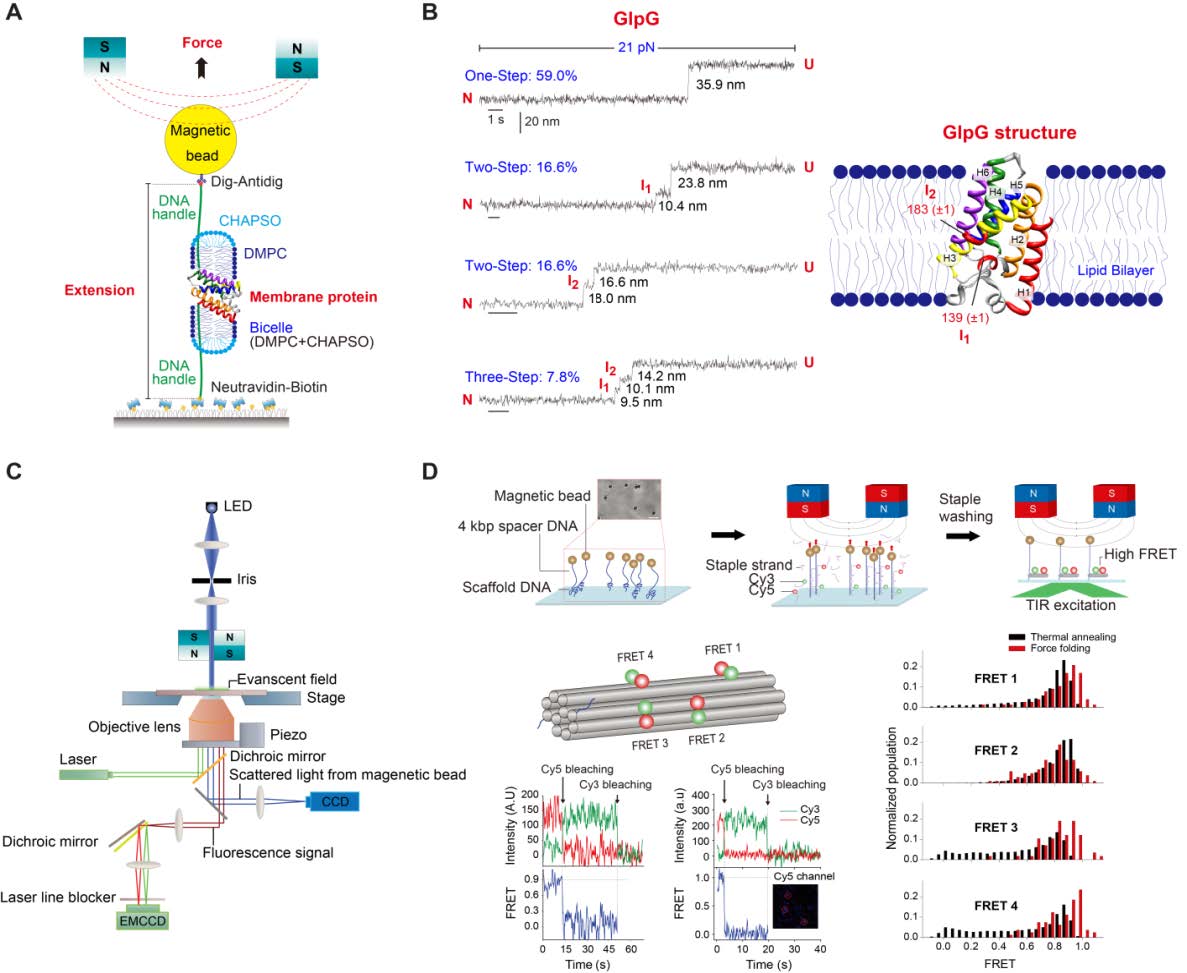
- Establish a setup that combines magnetic tweezers with single-molecule fluorescence. This technique will be used to apply force to membrane proteins to study conformational changes on individual molecules while assessing their functionalities with fluorescent probes.
- Develop a multi-color single-molecule FRET setup that will allow detection of synchronized or correlated motions among multiple domains.
- Develop a setup to measure single-molecule fluorescence with enhanced time resolution to resolve fast conformational changes
AIM 3: To further develop and enhance ensemble fluorescence techniques:
- Improvement of an LRET setup and make it available to members of the consortium to measure distances in functional membrane proteins.
- Develop a setup to measure nanosecond fluorophore lifetimes in the microsecond time scale combined with electrophysiology.
- Improve the fluorescence detection system with a new design of the photodetector-to-voltage transducer and develop a new more powerful acquisition system that will be used for all of the above setups.


