Effects of N-glycosylation on protein conformation and dynamics: Protein Data Bank analysis and molecular dynamics simulation study
By Hui Sun Lee, Yifei Qi and Wonpil Im.
Published in Sci Rep 2015 Mar 9;5:8926.
PMID: 25748215. PMCID: PMC4352867. Link to publication page.
Core Facility: Computational Modeling
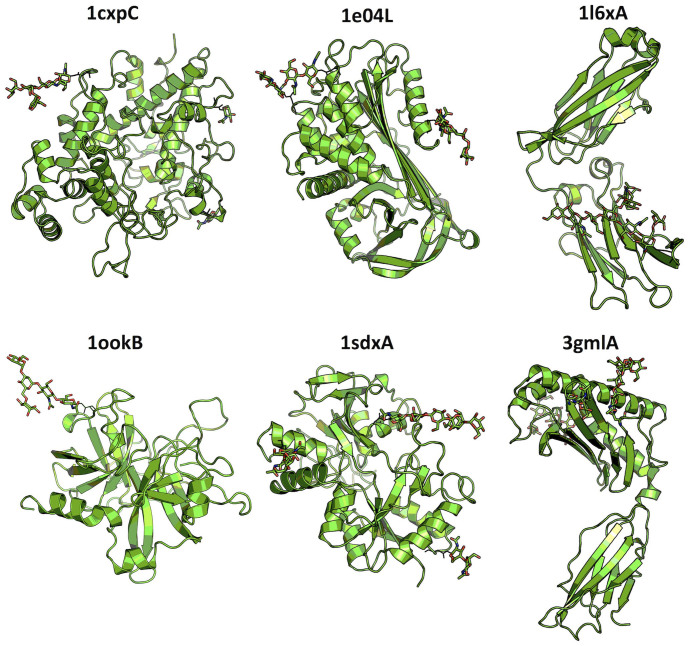
Figure 1.
The cartoon representation of six glycoproteins (PDB id + chain id) used for MD simulation study. The stick representation is used to display N-linked glycans.
Abstract
N-linked glycosylation is one of the most important, chemically complex, and ubiquitous post-translational modifications in all eukaryotes. The N-glycans that are covalently linked to proteins are involved in numerous biological processes. There is considerable interest in developments of general approaches to predict the structural consequences of site-specific glycosylation and to understand how these effects can be exploited in protein design with advantageous properties. In this study, the impacts of N-glycans on protein structure and dynamics are systematically investigated using an integrated computational approach of the Protein Data Bank structure analysis and atomistic molecular dynamics simulations of glycosylated and deglycosylated proteins. Our study reveals that N-glycosylation does not induce significant changes in protein structure, but decreases protein dynamics, likely leading to an increase in protein stability. Overall, these results suggest not only a common role of glycosylation in proteins, but also a need for certain proteins to be properly glycosylated to gain their intrinsic dynamic properties.


