Phage display selections for affinity reagents to membrane proteins in nanodiscs
By Pawel K. Dominik and Anthony A. Kossiakoff.
Published in Methods Enzymol. 2015;557:219-45. Epub 2015 Mar 24. PMID: 25950967. Link to publication page.
Core Facility: Synthetic Antigen Binder (SAB) Generation and Crystallography
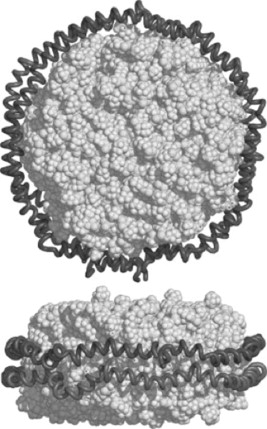
Figure 2. Structure of nanodisc. Nanodisc consists of lipid bilayer (white space filling model, here POPC are shown) and a double belt of membrane scaffold protein, MSP (gray ribbon). Adapted from Ritchie et al. (2009), with permission from Elsevier.
Abstract
Phage display selections generate high-affinity synthetic reagents that can be used as tools in structural characterization of membrane proteins. Currently, most selection protocols are performed with membrane protein targets in detergents. However, there are numerous technical issues associated with this, primarily that detergents are poor mimics of the native lipid environment. Here, we describe a set of protocols for phage display selection that involves reconstituting membrane proteins in nanodiscs, which are small discoidal particles consisting of lipids enclosed by membrane scaffold proteins. The nanodisc format enabled us to expand the capabilities of competitive and subtractive phage display selection steps, and generation of high-quality synthetic reagents for membrane proteins in native-like lipid environment.


