A gate-free pathway for substrate release from the inward-facing state of the Na(+)-galactose transporter
By Jing Li and Emad Tajkhorshid.
Published in Biochimica et Biophysica Acta (BBA) – Biomembranes 1818(2): 263-271 on February 2012.
PMID: 21978597. PMCID: PMC3253917. Link to Pubmed page.
Project: The Transport Cycle in Neurotransmitter Uptake Systems. Core Facility: Computational Modeling.
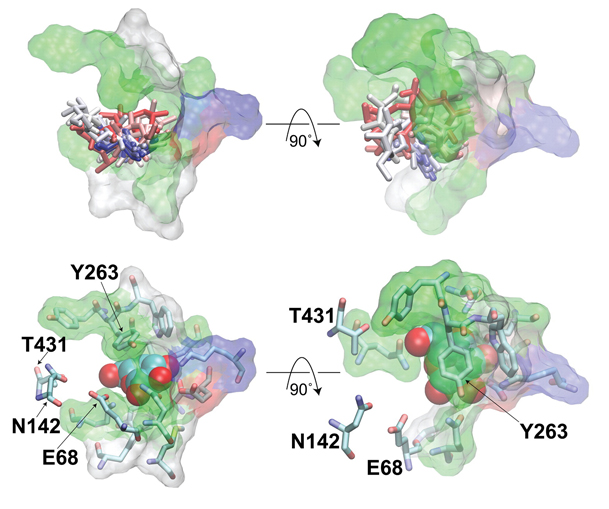
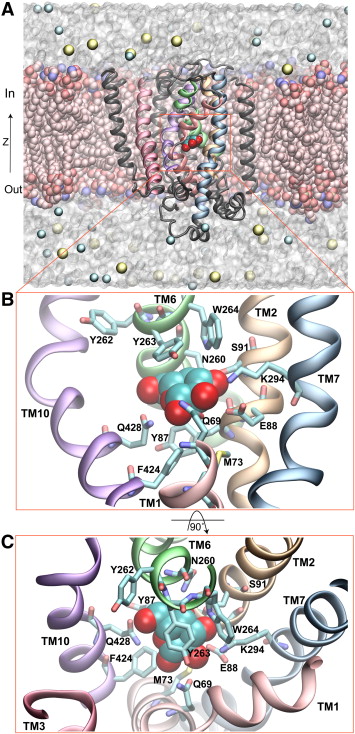
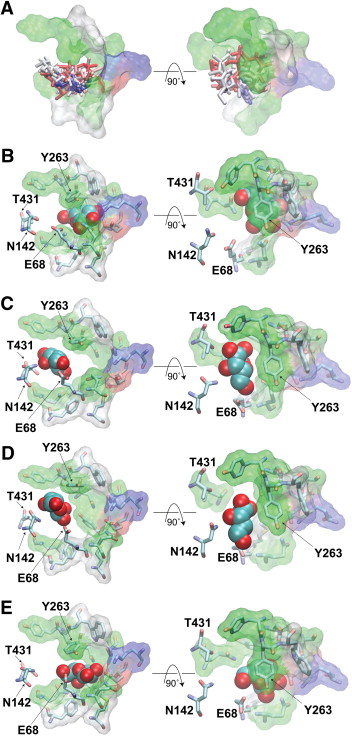
Spontaneous substrate unbinding in the equilibrium simulation. Residues in the substrate binding site are shown in overlaid stick and transparent surface representations. Residues that contact the substrate only during its unbinding from the original pocket, i.e., E68, N142, and T431, are only displayed as sticks.
Abstract
Employing molecular dynamics (MD) simulations, the pathway and mechanism of substrate unbinding from the inward-facing state of the Na+-coupled galactose transporter, vSGLT, have been investigated. During a 200-ns equilibrium simulation, repeated spontaneous unbinding events of the substrate from its binding site have been observed. In contrast to the previously proposed gating role of a tyrosine residue (Y263), the unbinding mechanism captured in the present equilibrium simulation does not rely on the displacement and/or rotation of this side chain. Rather, the unbinding involves an initial lateral displacement of the substrate out of the binding site which allows the substrate to completely emerge from the region covered by the side chain of Y263 without any noticeable conformational changes of the latter. Starting with the snapshots taken from this equilibrium simulation with the substrate outside the binding site, steered MD (SMD) simulations were then used to probe the translocation of the substrate along the remaining of the release pathway within the protein’s lumen and to characterize the nature of protein–substrate interactions involved in the process. Combining the results of the equilibrium and SMD simulations, we provide a description of the full translocation pathway for the substrate release from the binding site into the cytoplasm. Residues E68, N142, T431, and N267 facilitate the initial substrate’s displacement out of the binding site, while the translocation of the substrate along the remainder of the exit pathway formed between TM6 and TM8 is facilitated by H-bond interactions between the substrate and a series of conserved, polar residues (Y138, N267, R273, S365, S368, N371, S372, and T375). The observed molecular events indicate that no gating is required for the release of the substrate from the crystallographically captured structure of the inward-facing state of SGLT, suggesting that this conformation might represent an open, rather than occluded, state of the transporter. This article is part of a Special Issue entitled: Membrane protein structure and function.