CHARMM-GUI HMMM Builder for Membrane Simulations with the Highly Mobile Membrane-Mimetic Model
By Yifei Qi, Xi Cheng, Jumin Lee, Josh V. Vermaas, Taras V. Pogorelov, Emad Tajkhorshid, Soohyung Park, Jeffery B. Klauda, and Wonpil Im.
Published in Biophys J. 2015 Nov 17;109(10):2012-22. PMID: 26588561. PMCID: PMC4656882. Link to publication page.
Core: Computational Modeling Core.
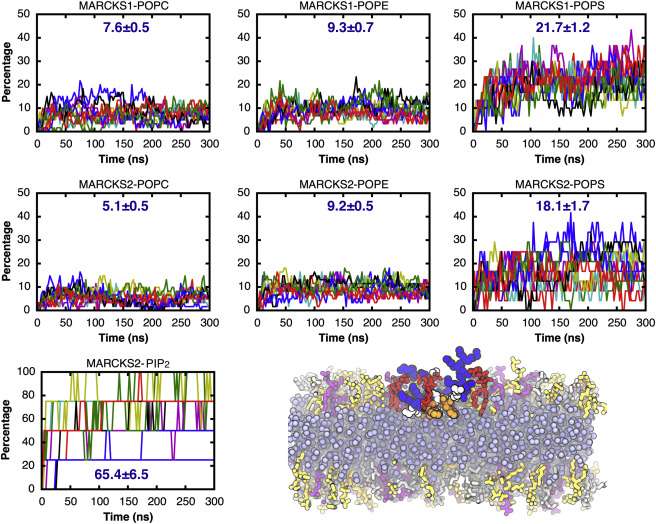
Figure 7. Percentages of lipids in contact with MARCKS-ED and a snapshot of the MARCKS2 system showing a MARCKS-ED in contact with three PIP2. A contact is counted when the minimal distance between the heavy atoms from a lipid and the peptide is <4.5 Å. Each colored line represents one peptide. The averages and standard errors of percentages from 150 ns to 300 ns are shown in blue numbers. In the snapshot, MARCKS-ED is shown in spheres with lysine colored in blue, phenylalanine and leucine in orange, and other residues in white; DCLE is shown in blue spheres, POPC in orange sticks, POPS in purple sticks, POPE in silver sticks, and PIP2 in red sticks. To see this figure in color, go online.
Abstract
Slow diffusion of the lipids in conventional all-atom simulations of membrane systems makes it difficult to sample large rearrangements of lipids and protein-lipid interactions. Recently, Tajkhorshid and co-workers developed the highly mobile membrane-mimetic (HMMM) model with accelerated lipid motion by replacing the lipid tails with small organic molecules. The HMMM model provides accelerated lipid diffusion by one to two orders of magnitude, and is particularly useful in studying membrane-protein associations. However, building an HMMM simulation system is not easy, as it requires sophisticated treatment of the lipid tails. In this study, we have developed CHARMM-GUI HMMM Builder (http://www.charmm-gui.org/input/hmmm) to provide users with ready-to-go input files for simulating HMMM membrane systems with/without proteins. Various lipid-only and protein-lipid systems are simulated to validate the qualities of the systems generated by HMMM Builder with focus on the basic properties and advantages of the HMMM model. HMMM Builder supports all lipid types available in CHARMM-GUI and also provides a module to convert back and forth between an HMMM membrane and a full-length membrane. We expect HMMM Builder to be a useful tool in studying membrane systems with enhanced lipid diffusion.


