Conformational coupling of the nucleotide-binding and the transmembrane domains in ABC transporters
By Po-Chao Wen and Emad Tajkhorshid
Published in Biophysical journal 101(3): 680-90 on August 3, 2011.
PMID: 21806936. PMCID: PMC3145290. Link to Pubmed page.
Project: Structural Dynamics of ABC Transporter
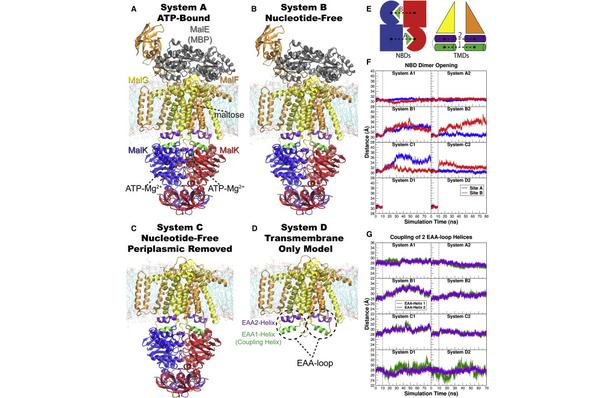
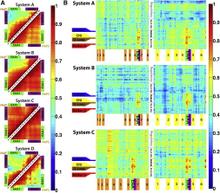
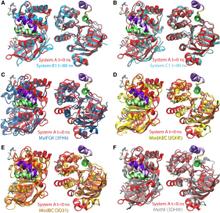
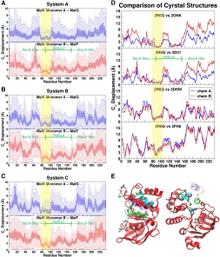
Figure 1. Overview of the equilibrium simulations. (A–D) Initial structure of the four simulation systems, showing proteins (in ribbons), bound substrates (including maltose and MgATP, shown as van der Waals spheres and labeled in panel A), and lipids (in line representations). The color scheme for this and all following figures is: MalK monomers in blue and red, MalF in orange, MalG in yellow, MalE in dark gray (labeled in panel A); the two EAA1 (the coupling helices) and EAA2 helices are highlighted in green and purple, respectively (labeled in panel D). (E) Schematic representation of the distances measured to quantify the NBD opening and the separation of the EAA helix pairs for panels F andG. Left: the NBD opening in each simulation system is measured as the center-of-mass distance between the helical subdomain of one MalK monomer (P88–E151, squares) and the RecA-like subdomain of the opposite MalK monomer (A2–Y87 and P152–G235, pac-man shaped); the bound nucleotides are shown as green triangles occupying the binding sites A and B. Right: the separation of the two sets of EAA helices are measured as the distances between their centers of masses (MalF:P396–G407 with MalG:D185–G196 as set 1, and MalF:F411–L422 with MalG:W200–S211 as set 2). (F) The conformational changes in the NBDs of each simulation system measured by the degree of NBD dimer opening. (G) The conformational changes of the EAA loops of the TMDs, measured as the separation of the two EAA helix pairs. The vertical dashed lines at 10 ns in panels F and G denote the time point at which all the simulations were branched out from their common parent equilibration simulation system.
Abstract
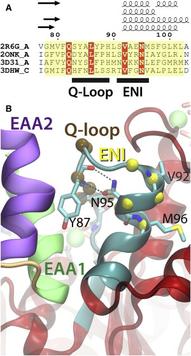
Basic architecture of ABC transporters includes two transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs). Although the transport process takes place in the TMDs, which provide the substrate translocation pathway across the cell membrane and control its accessibility between the two sides of the membrane, the energy required for the process is provided by conformational changes induced in the NBDs by binding and hydrolysis of ATP. Nucleotide-dependent conformational changes in the NBDs, therefore, need to be coupled to structural changes in the TMDs. Using molecular dynamics simulations, we have investigated the structural elements involved in the conformational coupling between the NBDs and the TMDs in the Escherichia coli maltose transporter, an ABC importer for which an intact structure is available both in inward-facing and outward-facing conformations. The prevailing model of coupling is primarily based on a single structural motif, known as the coupling helices, as the main structural element for the NBD-TMD coupling. Surprisingly, we find that in the absence of the NBDs the coupling helices can be conformationally decoupled from the rest of the TMDs, despite their covalent connection. That is, the structural integrity of the coupling helices and their tight coupling to the core of the TMDs rely on the contacts provided by the NBDs. Based on the conformational and dynamical analysis of the simulation trajectories, we propose that the core coupling elements in the maltose transporter involve contributions from several structural motifs located at the NBD-TMD interface, namely, the EAA loops from the TMDs, and the Q-loop and the ENI motifs from the NBDs. These three structural motifs in small ABC importers show a high degree of correlation in motion and mediate the necessary conformational coupling between the core of TMDs and the helical subdomains of NBDs. A comprehensive analysis of the structurally known ABC transporters shows a high degree of conservation of the identified 3-motif coupling elements only in the subfamily of small ABC importers, suggesting a distinct mode of NBD-TMD coupling from the other two major ABC transporter folds, namely large ABC importers and ABC exporters.