Generating conformation-specific synthetic antibodies to trap proteins in selected functional states
By Marcin Paduch, Akiko Koide, Serdar Uysal, Shahir S. Rizk, Shohei Koide, and Anthony A. Kossiakoff.
Published in Methods March 15, 2013;60(1):3-14. PMID: 23280336. PMCID: PMC3650108. Link to Pubmed page.
Core Facility: Synthetic Antigen Binder (SAB) Generation and Crystallography
Abstract
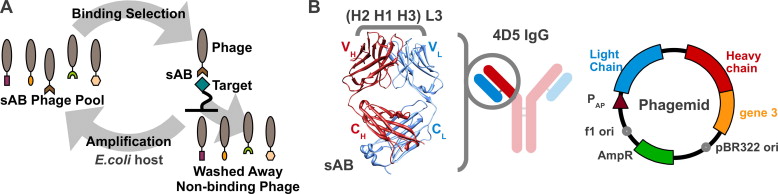
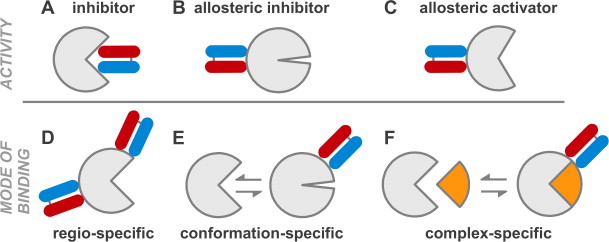
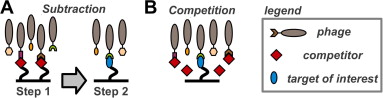
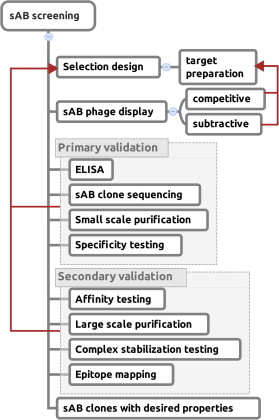
A set of phage display sorting strategies and validation methodologies are presented that are capable of producing high performance synthetic antibodies (sABs) with customized properties. Exquisite control of antigen and conditions during the phage display selection process can yield sABs that: (1) recognize conformational states, (2) target specific regions of the surface of a protein, (3) induce conformational changes, and (4) capture and stabilize multi-protein complexes. These unique capabilities open myriad opportunities to study complex macromolecular processes inaccessible to traditional affinity reagent technology. We present detailed protocols for de novo isolation of binders, as well as examples of downstream biophysical characterization. The methods described are generalizable and can be adapted to other in vitro direct evolution approaches based on yeast or mRNA display.