Structural basis of the alternating-access mechanism in a bile acid transporter
By Xiaoming Zhou, Elena J. Levin, Yaping Pan, Jason G. McCoy, Ruchika Sharma, Brian Kloss, Renato Bruni, Matthias Quick & Ming Zhou.
Published in Nature 505(7484): 569-73 on January 23, 2014. PMID: 24317697. PMC4142352. Link to publication page.
Project: Conformational Dynamics of a Glutamate Transporter Homologue.
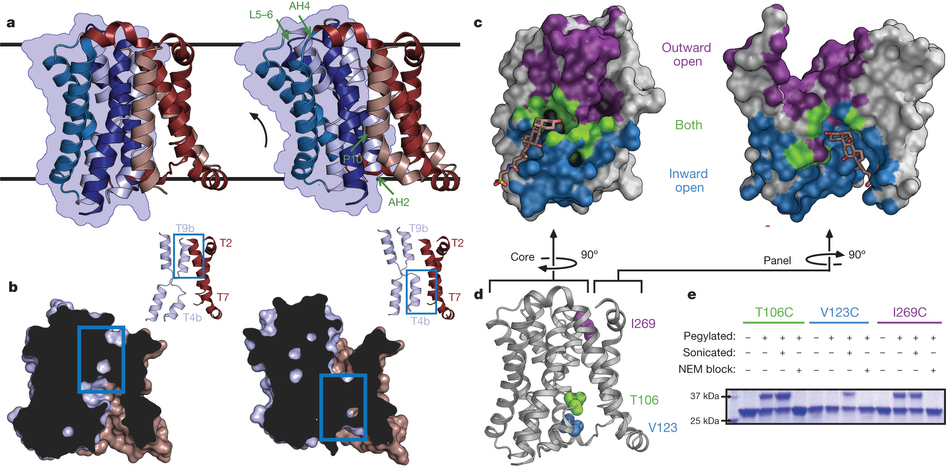
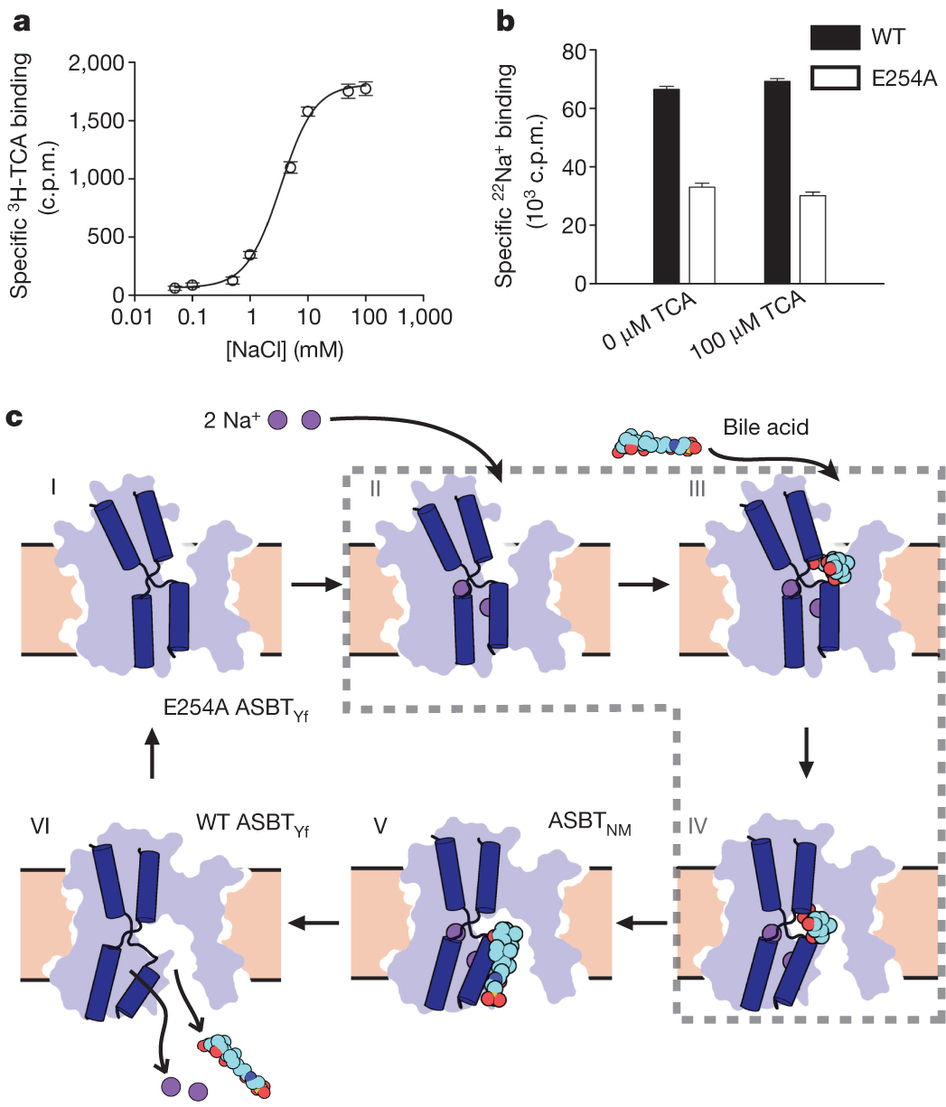
Figure 2. Structure and validation of the outward-open conformation of ASBTYf. a, The wild-type (left) and E254A (right) structures are shown side by side. Black lines correspond to the approximate position of the lipid bilayer inferred from the amphipathic helices. The core domain is marked with a blue silhouette, and regions acting as hinges in the conformational change between the two structures are marked with green arrows in the E254A structure. b, Cutaway view of the surfaces of the wild-type (left) and E254A (right) structures, showing the intracellular and extracellular cavities. Insets show key helices forming the interface between the panel and core domain; blue rectangles show the location of the interdomain interface in both structures. c, Surface representations of the core (left) and panel (right) domains of the wild-type ASBTYf structure. The sides of both domains facing the central cavity are coloured according to whether they are accessible to the cytoplasm in the wild-type structure (blue), accessible to the periplasm in the E254A structure (violet), or are solvent accessible in both conformations (green). A stick representation of TCA marks the location of the substrate in the ASBTNM structure. d, Cartoon representation of the wild-type ASBTYf structure with the locations of Thr 106, Val 123 and Ile 269 marked with spheres. e, SDS–polyacrylamide gel electrophoresis (SDS–PAGE) gel showing results of pegylation experiments for the three ASBTYf cysteine mutants. Pluses and minuses mark whether or not samples were incubated with mPEG-Mal-5K, were sonicated before pegylation to rupture the cell membranes, or were incubated with N-ethylmaleimide (NEM) before pegylation to prevent further modification of the cysteine residues.
Abstract
Bile acids are synthesized from cholesterol in hepatocytes and secreted through the biliary tract into the small intestine, where they aid in absorption of lipids and fat-soluble vitamins. Through a process known as enterohepatic recirculation, more than 90% of secreted bile acids are then retrieved from the intestine and returned to the liver for resecretion. In humans, there are two Na(+)-dependent bile acid transporters involved in enterohepatic recirculation, the Na(+)-taurocholate co-transporting polypeptide (NTCP; also known as SLC10A1) expressed in hepatocytes, and the apical sodium-dependent bile acid transporter (ASBT; also known as SLC10A2) expressed on enterocytes in the terminal ileum. In recent years, ASBT has attracted much interest as a potential drug target for treatment of hypercholesterolaemia, because inhibition of ASBT reduces reabsorption of bile acids, thus increasing bile acid synthesis and consequently cholesterol consumption. However, a lack of three-dimensional structures of bile acid transporters hampers our ability to understand the molecular mechanisms of substrate selectivity and transport, and to interpret the wealth of existing functional data. The crystal structure of an ASBT homologue from Neisseria meningitidis (ASBT(NM)) in detergent was reported recently, showing the protein in an inward-open conformation bound to two Na(+) and a taurocholic acid. However, the structural changes that bring bile acid and Na(+) across the membrane are difficult to infer from a single structure. To understand the structural changes associated with the coupled transport of Na(+) and bile acids, here we solved two structures of an ASBT homologue from Yersinia frederiksenii (ASBTYf) in a lipid environment, which reveal that a large rigid-body rotation of a substrate-binding domain gives the conserved ‘crossover’ region, where two discontinuous helices cross each other, alternating accessibility from either side of the cell membrane. This result has implications for the location and orientation of the bile acid during transport, as well as for the translocation pathway for Na(+).

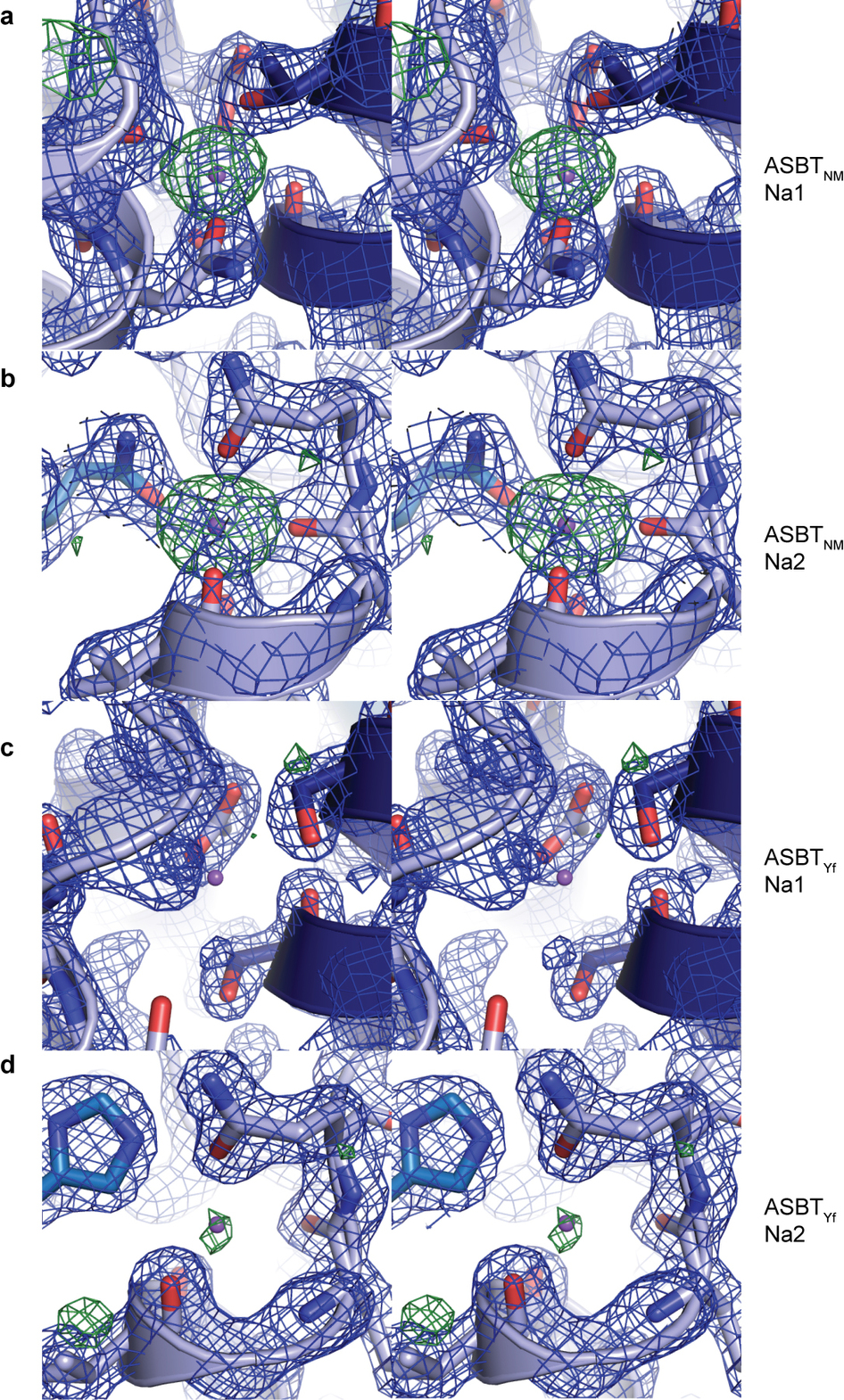
![Figure 1. Function and crystal structure of ASBTYf. a, Time course of uptake of 3H-TCA into empty or ASBTYf-containing proteoliposomes in the presence of 100 mM external NaCl or choline chloride. b, Uptake of 3H-TCA 30 s after addition to ASBTYf-containing proteoliposomes in the presence of 100 mM external NaCl or choline chloride, as a function of the external 3H-TCA concentration. c, Cartoon representation of the ASBTYf structure shown from two perpendicular directions in the plane of the membrane with the periplasm on top (left and middle), and from the extracellular side (right). The transmembrane helices are coloured in pseudosymmetry-related pairs. d, A cutaway surface representation of ASBTYf showing the locations of the discontinuous helices TM4 and TM9 relative to the intracellular cavity. Locations of the Na+-binding sites in the previously reported ASBTNM structure are marked with dotted circles. Inset on the right shows a magnified view of residues that coordinate Na+ in the ASBTNM structure. e, Alignment of the discontinuous helices TM4 and TM9 in the ASBTYf (light blue) and ASBTNM (dark blue) structures. The partly unwound region of TM4b is marked with a black arrow. f, Na+-binding kinetics of ASBTYf and the Na+-site mutants. Equilibrium binding of 0.95 µM [22Na]Cl (5.92 Ci mmol−1) to 250 ng of wild-type (WT), E254A or Q258A ASBTYf was measured with the SPA in the presence of increasing NaCl concentrations ranging from 0–100 mM. Isotopic replacement of 22Na+ was plotted as a function of the concentration of non-labelled NaCl. The means of triplicate measurements ± standard error of the mean (s.e.m.) were subjected to nonlinear regression fitting in Prism 5 (GraphPad) for panels a, b and f.](/site-media/images/publications/2013/Zhou2-2013-figure1.jpg)