Transport dynamics in a glutamate transporter homologue
By Nurunisa Akyuz, Roger B. Altman, Scott C. Blanchard, and Olga Boudker.
Published in Nature June 23, 2013;502(7469). PMID: 23792560. PMCID: PMC3829612. Link to publication page.
Project: Conformational Dynamics of a Glutamate Transporter Homologue
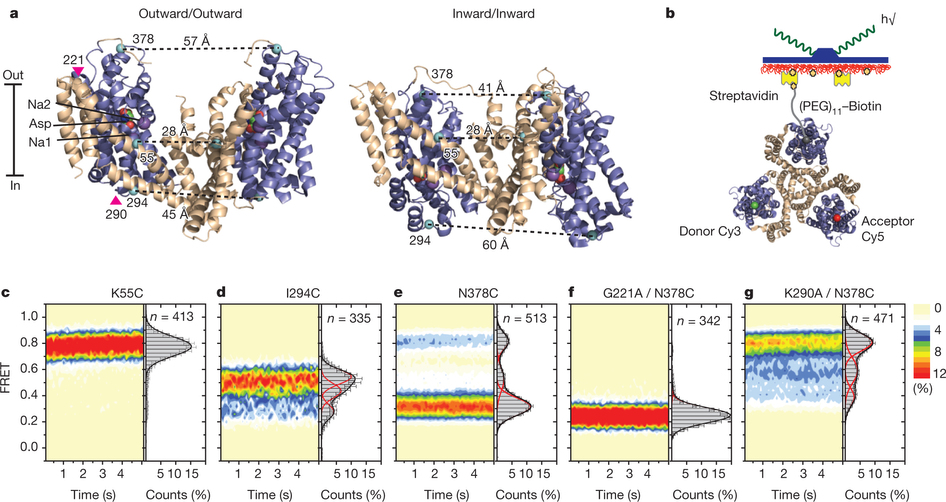
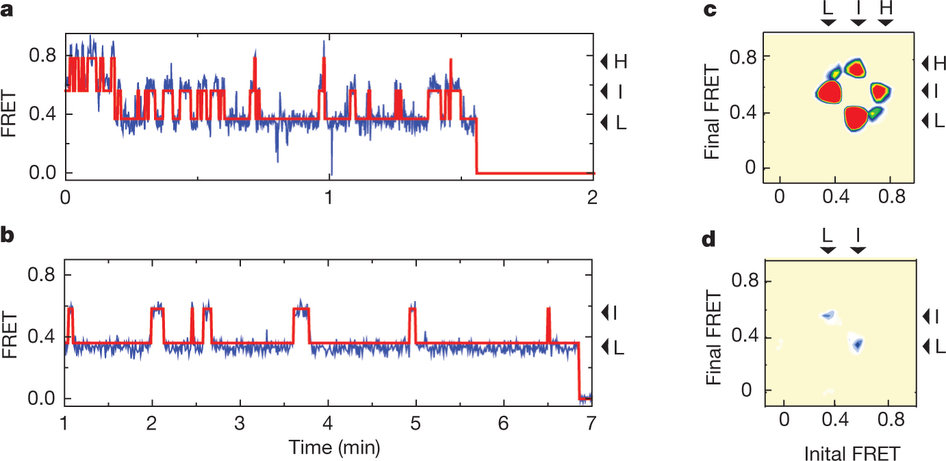
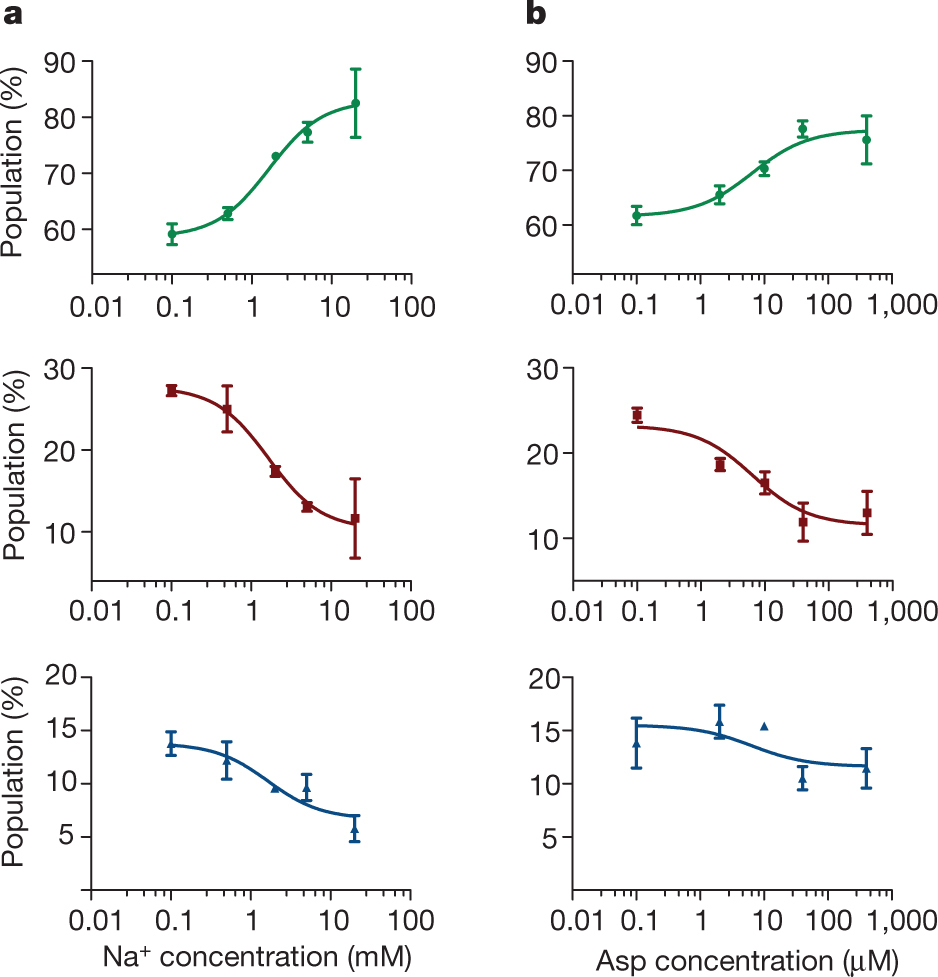
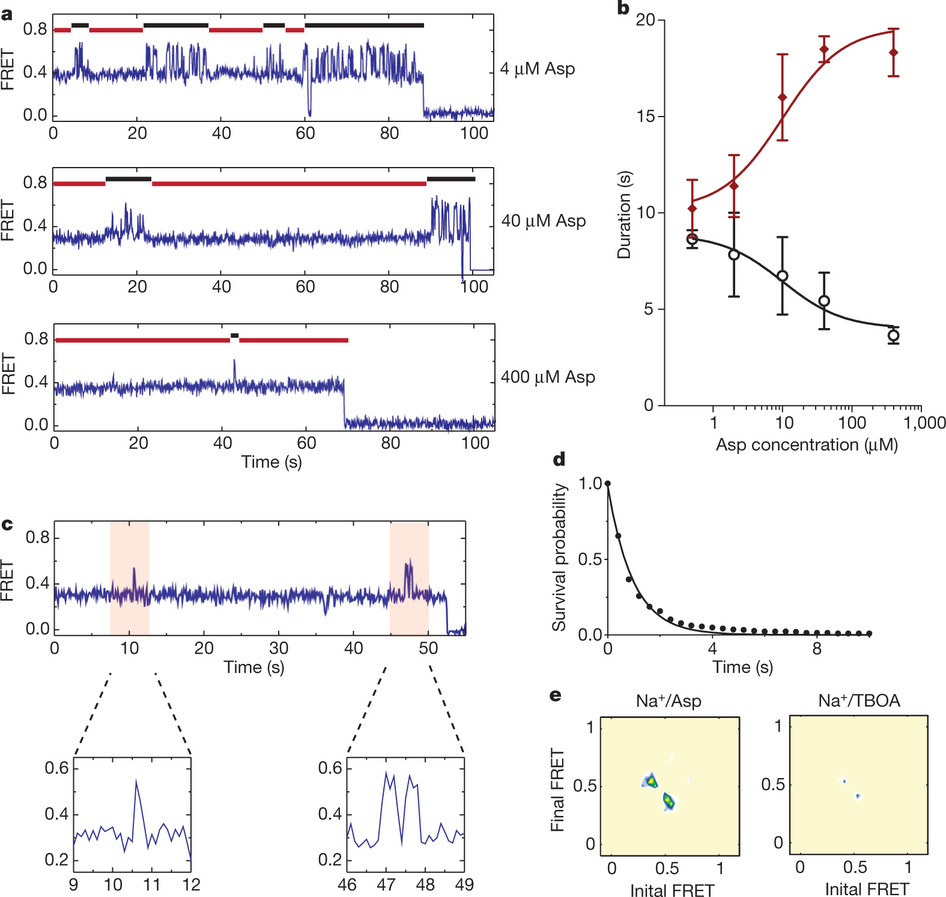
Figure 1: FRET efficiency changes reflect relative orientations of the transport domains.a, GltPh protomer pairs in symmetrical outward- and inward-facing states viewed within the membrane plane. Trimerization and transport domains are coloured wheat and blue, respectively. Bound Asp and Na+ ions are emphasized as spheres and coloured by atom type. Introduced cysteines are highlighted in cyan with inter-protomer distances above the dotted lines. Magenta arrows mark sites of mutations altering state distributions. b, Labelling and surface-immobilization strategies. c–g, FRET efficiency population histograms for Asp/Na+-bound transporters. Introduced mutations are indicated above the panels. The number of molecules analysed (n) is shown. Population contour plots (left) are colour-coded from tan (lowest) to red (highest population) with the colour scale shown beside the graphs. The cumulative population histogram (right) displays the time-averaged values and standard deviations. The solid black lines are fits to the sums of individual Gaussian functions (red lines).
Abstract
Glutamate transporters are integral membrane proteins that catalyse neurotransmitter uptake from the synaptic cleft into the cytoplasm of glial cells and neurons. Their mechanism of action involves transitions between extracellular (outward)-facing and intracellular (inward)-facing conformations, whereby substrate binding sites become accessible to either side of the membrane. This process has been proposed to entail transmembrane movements of three discrete transport domains within a trimeric scaffold. Using single-molecule fluorescence resonance energy transfer (smFRET) imaging, we have directly observed large-scale transport domain movements in a bacterial homologue of glutamate transporters. We find that individual transport domains alternate between periods of quiescence and periods of rapid transitions, reminiscent of bursting patterns first recorded in single ion channels using patch-clamp methods. We propose that the switch to the dynamic mode in glutamate transporters is due to separation of the transport domain from the trimeric scaffold, which precedes domain movements across the bilayer. This spontaneous dislodging of the substrate-loaded transport domain is approximately 100-fold slower than subsequent transmembrane movements and may be rate determining in the transport cycle.