Ouabain binding site in a functioning Na+/K+ ATPase
By Walter Sandtner, Bernhard Egwolf, Fatemeh Khalili-Araghi, Jorge E Sanchez-Rodriguez, Benoît Roux, Francisco Bezanilla, and Miguel Holmgren
Published in The journal of biological chemistry 286(44): 38177-83 on September 12, 2011.
PMID: 21911500. PMCID: PMC3207469. Link to Pubmed page.
Project: Conformational Transitions in P-class ATPases
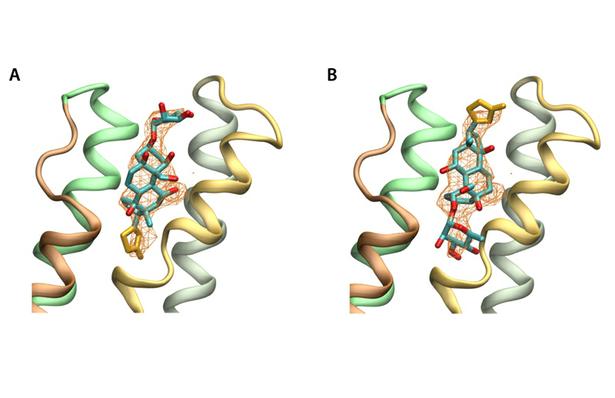
Figure 5. Orientation of ouabain in the alpha subunit based on structural factors of the PDB(3A3Y). Simulated annealed omit maps are shown at 2.5 sigma, with ouabain in the published (A) orreoriented 180º (B). The omit maps are compatible with both possible orientations of ouabain.
Abstract
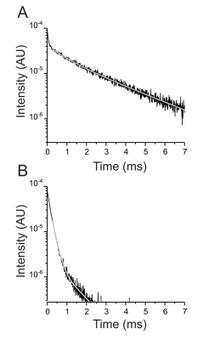
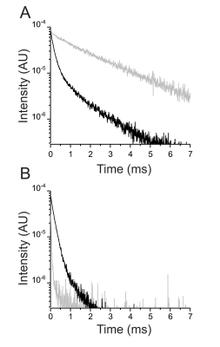
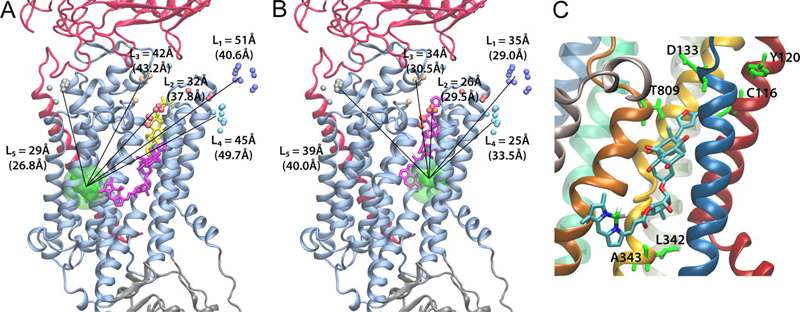
The Na+/K+ ATPase is an almost ubiquitous integral membrane protein within the animal kingdom. It is also the selective target for cardiotonic derivatives, widely prescribed inhibitors for patients with heart failure. Functional studies revealed that ouabain-sensitive residues distributed widely throughout the primary sequence of the protein. Recently, structural work has brought some consensus to the functional observations. Here, we use a spectroscopic approach to estimate distances between a fluorescent ouabain and a lanthanide binding tag (LBT), which was introduced at five different positions in the Na+/K+ ATPase sequence. These five normally functional LBT-Na+/K+ ATPase constructs were expressed in the cell membrane of Xenopus laevis oocytes, operating under physiological internal and external ion conditions. The spectroscopic data suggest two mutually exclusive distances between the LBT and the fluorescent ouabain. From the estimated distances and using homology models of the LBT-Na+/K+ ATPase constructs, approximate ouabain positions could be determined. Our results suggest that ouabain binds at two sites along the ion permeation pathway of the Na+/K+ ATPase. The external site (low apparent affinity) occupies the same region as previous structural findings. The high apparent affinity site is, however, slightly deeper towards the intracellular end of the protein. Interestingly, in both cases the lactone ring faces outward. We propose a sequential ouabain binding mechanism that is consistent with all functional and structural studies.