Summary of Progress
The project got underway 10/1/2010. In the first year of activity, the experimental focus was to select and develop the expression platform. In parallel, computational analysis was initiated to characterize the dynamic modes of apo Pgp and simultaneously select sites for incorporation of spin label probes. The summary describes progress on the specific aims. The synergy with cores, which underlies the rationale for the consortium, is emphasized in projects described as research highlights (RH). The pending publication of the first crystal structure of an ABC heterodimer spurred us to initiate spin labeling studies of this subclass of transporters (RH #1). Synthesis of spin labeled Pgp substrate analogs is providing a new approach for investigation of transport (RH #3). Alternative approaches to Pgp expression are being pursued (RH #3) and novel computational tools (RH #4) are being developed to analyze distance distributions between spin labels. Many of these methodological developments will be critical to the success of this project and can be applied in other bridges and by the membrane protein community at large.
Construction and Expression of cysteine less human Mdr1 in insect cells.
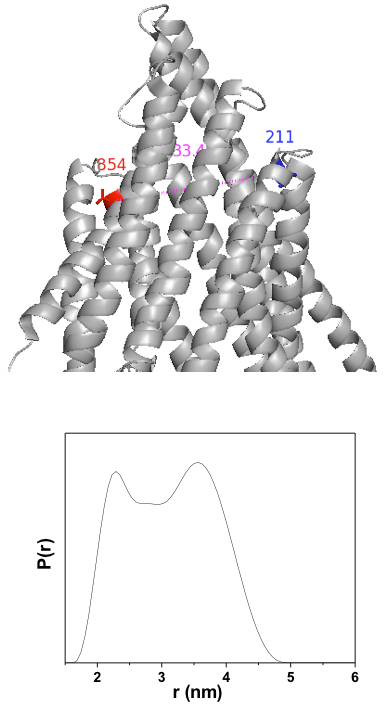
Figure 1. Close up view of sites 215/858 (human sequence) in the apo Pgp crystal structure. Lower panel: DEER distance distribution showing a clear bimodal shape suggesting equilibrium between multiple conformations.
Click to enlarge.In collaboration with the Membrane Protein Expression / Purification Core, we compared Pgp expression in Saccharomyces cerevisiae to that in insect cells infected with baculovirus. The results suggested the higher eukaryotic system is better suited for production of the mammalian transporter at the scales required for EPR spectroscopy. While yeast cells are easier and less expensive to grow, the difficulties in solubilizing the protein from the ergosterol-containing yeast membranes is a distinct disadvantage compared to insect cells. The Mchaourab lab cloned human P-glycoprotein gene into pFastBac HT A (Invitrogen). There is a 28 residues linker before the start of the P-gp gene that includes a 6x His tag. To generate P-gp Cless, all seven cysteines (C137, C431, C717, C956, C1074, C1125, and C1227) were mutated to alanines. Figure 1 shows the expression and purification of human Pgp detected by western blotting.
The Protein Core has further developed the expression system of the cysteine-less human P-glycoprotein. The original cysteine-less baculovirus construct produced in the Mchaourab lab was tested in different insect cell lines. The highest expression system was grown at large scale and the protein purified via its (His)6 affinity tag. Based on results from molecular dynamics simulations (see below), five double.
DEER analysis of apo Pgp structure.
We have succeeded in obtaining DEER data for the double mutant 215/858 (human sequence). Figure 1 shows that the distance distribution for this double mutant is bimodal suggesting equilibrium fluctuations at the extracellular side of the transporter. The long distance component is consistent with apo Pgp crystal structure corrected for the projection of the spin label side chain. The shorter component is suggestive of a more open conformation. Although preliminary, this data highlights the unique insight obtained from spectroscopic analysis of this transporter. Five double mutants have been constructed and are now in production stage. An initial mapping of distance changes upon ATP hydrolysis will be performed in the next year of activity.
Research Highlights
Click to find out more about each research highlight (RH).
Gustot Smriti, Hassane Mchaourab
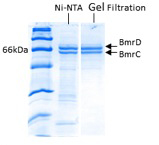
Figure 2. Typical SDS-PAGE detection of purified BmrC/D.
For ABC transporters, dimerization of conserved motifs in the nucleotide binding domains (NBDs) is a critical step in ATP binding and hydrolysis. A subclass of the bacterial ABC transporter superfamily is assembled from two distinct gene products, i.e. they form a heterodimer. In some ABC heterodimers such as BmrC/BmrD from Bacillus subtilis, the two NBDs are sequence asymmetric suggesting that nucleotide binding and hydrolysis occurs in only one of the two ATP binding sites. BmrC contains a so called “degenerate site”, characterized by mutation of a glutamate to aspartate in the Walker B motif. Asymmetric hydrolysis has been invoked in the transport cycle of human transporter such as the transporter associated with antigen processing (TAP) and the cystic fibrosis conductance regulator (CFTR). Thus it is imperative to understand the conformational cycle of these asymmetric transporters. In addition, a structure of a thermophilic homolog was presented at the 2011 multidrug efflux Gordon Conference and it should be published shortly. Thus this class of transporter provides an ideal new target to add to the consortium list. We have initiated site-directed spin labeling studies of the conformational cycle of BmrC/BmrD. For this purpose, we have procured the expression vector of BmrC/BmrD. As shown in Figure 2, heterodimer can be expressed and purified by nickel affinity followed by size exclusion chromatography. We have also replaced the three native cysteines. Future studies will measure distances between the different domains of BmrC/D in order to assess the magnitude of ATP driven conformational changes.
Louis Luk, Steve Kent, Hassane S. Mchaourab
In collaboration with the chemical synthesis core
ABC transporters, including Pgp, bind and transport a spectrum of molecules with diverse chemical structures. Historically, binding has been detected through enhancement of ATPase activity. In some cases, changes in the fluorescence intensity of optically active substrates have also been used to characterize binding. For both approaches, there are intrinsic limitations that preclude direct detection of binding. Specifically, fluorescence is influenced by partitioning of the substrate in lipid bilayers. In addition to its utility for binding analysis, a spin labeled substrate will be ideal for distance measurements to spin labels site specifically incorporated into Pgp. Therefore, we have initiated a collaboration with the chemistry core to synthesize spin labeled analogs of selected Pgp substrates. So far, we have synthesized and tested spin labeled doxorubicin. This molecule binds Pgp and its bacterial homologs such as MsbA. The binding is reduced by addition of ADP and vanadate. Interestingly, this probe also binds other multidrug transporters and may prove of general utility in the field.
Hanane A. Koteiche, Volker Dötsch, Hassane S. Mchaourab
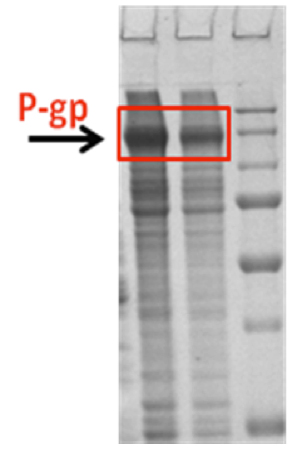
Figure 3. Cell-free expression of Pgp.
Click to enlarge.In collaboration with the membrane protein expression / purification core
Cell-free expression provides an alternative platform for expression of membrane proteins. Advantages of this approach include the possibility of adding agents that support expression and stabilize the protein and the direct synthesis of the protein into detergent micelles or lipid vesicles. In collaboration with the protein core, we have implemented cell-free expression protocols for Pgp. Drs. Mchaourab and Koteiche (Mchaourab Lab) visited the Dötsch laboratory at the University of Frankfurt for two weeks in order to gain hands on experience in the preparation of reagents and optimization of expression conditions. As shown in the SDS-PAGE of Figure 3 (see RH #2), we were able to obtain robust expression of Pgp in the precipitate mode; i.e. in the absence of detergents. The expression levels approach 1 mg/ml which is far superior to any recombinant method. The challenge will be to express Pgp at similar levels in the detergent mode in order to keep it soluble and then carry out detergent exchange to maximize its activity. The plan will be to use drug-stimulated ATPase activity to functionally characterize Pgp expressed in a cell-free format.

Figure 4. Structure of T4L highlighting representative pairs used for distance measurements between spin labels. σ values calculated from experimental distance distributions from T4L are shown as a histogram binned at intervals of 0.5Å.
Click to enlarge.Richard Stein, Shahid Islam, Hassane S. Mchaourab, Benoît Roux
In collaboration with the computational modeling core
Recent advances in pulsed EPR spectroscopy enables distance measurement between pairs spin labels in the range of 20-60Å. Model-free analysis of double electron resonance (DEER) experiments provides the probability distribution of the distance between two spin labels (histogram of distances). The approach is very promising to monitor large-scale conformational changes in ABC transporters. One of the aims of the computational core is help establish robust computational framework to translate such experimental information into structural restraints.
For this purpose, ~60 pairs of nitroxide spin labels were introduced at surface sites in T4 lysozyme (T4L), focusing on the helical C-terminal domain and avoiding regions of the protein affected by the hinge bending motion in solution. Each of the resulting distance distributions was measured and parametrized by the weighted average rav and the width s. The set of measurements on T4L provides a unique training ground to develop and validate a new algorithm for the analysis of DEER data whereby the ~60 histogram of distances are used as restraints in molecular dynamics simulations.
Po-Chao Wen and Emad Tajkhorshid
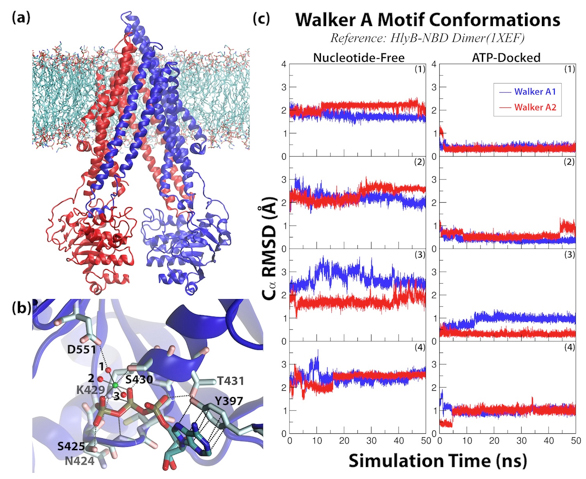
Figure 5. Overview of PgP simulations. (a) The simulation system. (b) Close-up view of the Mg
2+-ATP-docked nucleotide binding site. The Mg
2+ is drawn as a green sphere and its coordinating water in red. (c) RMSD of the Walker A motif during the simulations.
Click to enlarge.In order to characterize the dynamics of PgP, the protein has been modeled in an explicit membrane environment and simulated using equilibrium simulations. The results show that, surprisingly, Pgp is capable of adopting an unusually wide range of conformations even in its nucleotide-free form, as highlighted by the degree of variation in the distance between the two nucleotide-binding domains (NBDs) over a range of ~20A, exhibiting conformations both more open and closed than the crystal structure. In addition, we have been able to successfully dock Mg-ATP in the Walker A motif, and generate an ATP-bound state that remains stable throughout the simulations. The presence of the nucleotide shifts the distribution of NBD conformations toward a more closed form, however, the protein continues to exhibit large degree of fluctuations. These results along with the reported high NBD fluctuations measured in several other ABC exporters, suggest that a high degree of fluctuation might be a common feature to the transport mechanisms in ABC exporters, which is fundamentally different from ABC importers. During these simulations, we have also captured events of lipid penetration directly from the bilayer into the drug binding pocket of PgP, as well as opening of the TMDs from the extracellular side. The location of the protruding lipid tail, together with the extracellular opening in the TMDs provide probable pathways for drug entrance and exit during the transport cycle.
Wenxun Gan, Mahmoud Moradi, and Emad Tajkhorshid
As a PgP homolog whose structure has been captured in additional states, we have also taken advantage of the structure of MsbA to characterize the structural transitions involved in interconversion of different states of ABC exporters. We have performed MD simulations together with a combination of several computational methods. Starting from the recently solved X-ray crystal structure of MsbA in the OF state, we first generate high-resolution all-atom models for the two IF states by using targeted MD simulations. We then define two collective variables to describe the relative motion of the two transmembrane domains (TMDs) and nucleotide binding domains (NBDs), respectively. Steered MD simulations along the collective variables have been performed to induce the conformational changes between the three states of MsbA. Assessing the energetics associated with the induced transitions, approximated by calculating the work involved in going from one state to another, suggests that the OF-to-IF conformational transition follows two steps: the two TMDs close first, and then the two NBDs open, as opposed to the pathway observed from initial targeted MD simulations. Taken together, these results not only provide a better understanding of the functionality of ABC transporters, but also help define a general mechanism for membrane transport process. The method used in these simulations is developed within our activity in Computational Modeling Core and specifically for this project.
Po-Chao Wen, Christopher Mayne, and Emad Tajkhorshid
One of the main goals in this Bridge Project is to characterize the binding site(s) for various drugs in PgP and other drug exporters, and how ligand binding to the protein might affect the energy landscape of the different states of the protein. In order to do so, we need to develop compatible force field parameters for the drugs that are used experimentally to study the activity of the transporter. This step is a tedious task in computational projects. Taking advantage of the Computational Modeling Core, we have developed a set of force field parameters for rhodamine and verapamil, and have completed initial steps of parameterization for doxorubicin. Furthermore, the membrane partitioning of verapamil, which will determine the pathway through which it binds to the binding pocket/cavity in the transporter, has been also simulated using a novel membrane representation developed in the group.
Wenxun Gan and Emad Tajkhorshid

Figure 6.
Click to enlarge.NorM is a member of the MATE (multidrug and toxic compound extrusion) transporter family, which use either H+ or Na+ gradients to translocate their substrates across the cellular membrane. Similar to PgP, NorM is an exporter that acts on various drugs, and therefore, its mechanistic studies are relevant to our understanding of the nature of conformational changes involved in the export process. Starting on a recent X-ray crystal structure of NorM in the outward-facing (OF) conformation, we have performed MD simulations with explicit solvent and lipids to study the coupling of cation binding, conformational transitions, and substrate export pathways in NorM transport cycle. Specifically, we want to investigate the atomistic details of the OF-to-IF and IF-to-OF conformational changes, the mechanism by which Na+ binding to NorM might affect the conformational equilibrium of NorM, and binding sites and translocation pathways of small molecules (drugs) in NorM. We have performed extended equilibrium MD simulations of NorM, with and without Na+ bound (200 ns each). In the Na+-bound simulation, the three transmembrane helices surrounding the Na+ binding site show significantly larger motion compared to the apo simulations, suggesting that NorM has a tendency to undergo the OF-to-IF conformational change with Na+ bound. In order to study the conformational transitions between the OF and IF states, a key step will be to model the unknown IF state first. Currently, we’re working on initial building and refining the IF model by using biased simulations performed in the space of specific collective variables. We have also generated a set of parameters for the small drug molecule, doxorubicin (generated by GAFF/Antechamber; and being parameterized for CHARMM) which will be docked to NorM different states and its translocation and how it affects the protein will be studied with steered MD simulations.
 Figure 1. Close up view of sites 215/858 (human sequence) in the apo Pgp crystal structure. Lower panel: DEER distance distribution showing a clear bimodal shape suggesting equilibrium between multiple conformations. Click to enlarge.
Figure 1. Close up view of sites 215/858 (human sequence) in the apo Pgp crystal structure. Lower panel: DEER distance distribution showing a clear bimodal shape suggesting equilibrium between multiple conformations. Click to enlarge.






