The membrane environment is a key determinant of function and/or organization of membrane proteins. It is essential for the activities of the Consortium to have access to quantitative information about the characteristics and energetics of membrane-protein interactions. For multi-segment transmembrane proteins in particular, such characterization is complicated by the different hydrophobic thicknesses of the component transmembrane segments and the radially non-uniform hydrophobic interface created between the membrane and the protein. A hybrid Continuum-Molecular Dynamics (CTMD) approach to compute membrane deformations profiles around multi-segment proteins and corresponding energetics was recently published [Sayan Mondal, George Khelashvili, Jufang Shan, Olaf Andersen, Harel Weinstein (2011), Biophysical J (101): 2092-2101; link]. This stand-alone application implements the CTMD approach.
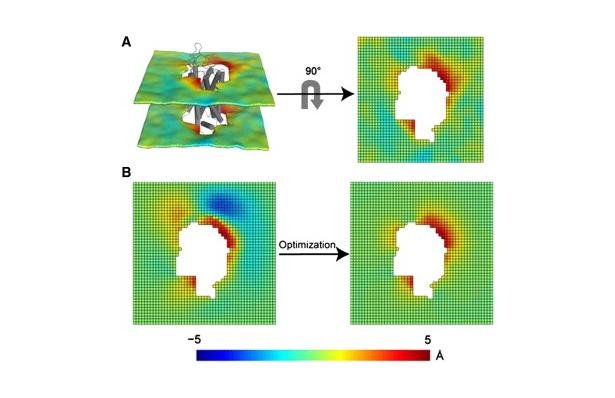 Protocol for the 3D-CTMD approach, illustrated for rhodopsin in a diC14:1PC lipid bilayer (Mondal et al., BJ 2011). Click to enlarge and learn more.
Protocol for the 3D-CTMD approach, illustrated for rhodopsin in a diC14:1PC lipid bilayer (Mondal et al., BJ 2011). Click to enlarge and learn more.The CTMDapp software calculates the deformation profiles of the bilayer and the free energy cost of the membrane deformation around multi-segment transmembrane proteins, taking into account the radially non-uniform hydrophobic surface of the protein. As the primary input it requires a molecular dynamics trajectory of a multi-segment, transmembrane protein embedded in a bilayer.
To allow for calculations without a molecular dynamics input, the CTMDapp software also implements a simplified version of the CTMD method that can assess the radial asymmetry of the membrane-protein interface for a particular protein structure at an approximate level.
Methodological details can be found in the original paper (Mondal et al., BJ 2011). In addition, the application is documented with brief usage notes at every step and generates diagnostic intermediate output.
Download the software:
- Windows: http://memprotein.org/software/CTMDapp_windows_pkg.exe
- Mac: http://memprotein.org/software/CTMDapp_mac_pkg.zip
- Unix: http://memprotein.org/software/CTMDapp_unix_pkg.zip
Instructions:
For the Windows version, clicking on the downloadable .exe file will directly launch the Matlab Component Runtime (MCR) installation process and extract the app executable. For the Mac version, the MCR must be installed first by opening MCRinstaller.dmg in the folder. This is a one-time installation, after which CTMDapp can be launched. The Unix version requires an interface like Xwin along with a script setting proper paths.
The software was written by Sayan Mondal (mos2013@med.cornell.edu) in the Harel Weinstein (haw2002@med.cornell.edu) lab at Weill Cornell Medical College, Cornell University. Please feel free to contact us with questions and suggestions about the software.







