The MPSDC had a strong showing at the Biophysical Society’s 56th Annual Meeting this year, with members collaborating in a total of 72 posters and presentations taking place in various subgroups, platforms, symposia, mini-symposia, and workshops. Now, we are proud to announce that one of our posters was selected as one of the thirteen 2012 Student Research Achievement Award Winners.
Thomas Chew, the first author of the poster, is an undergraduate student at the University of California, San Diego. He has performed summer research projects in the laboratory of Merritt Maduke at Stanford University.
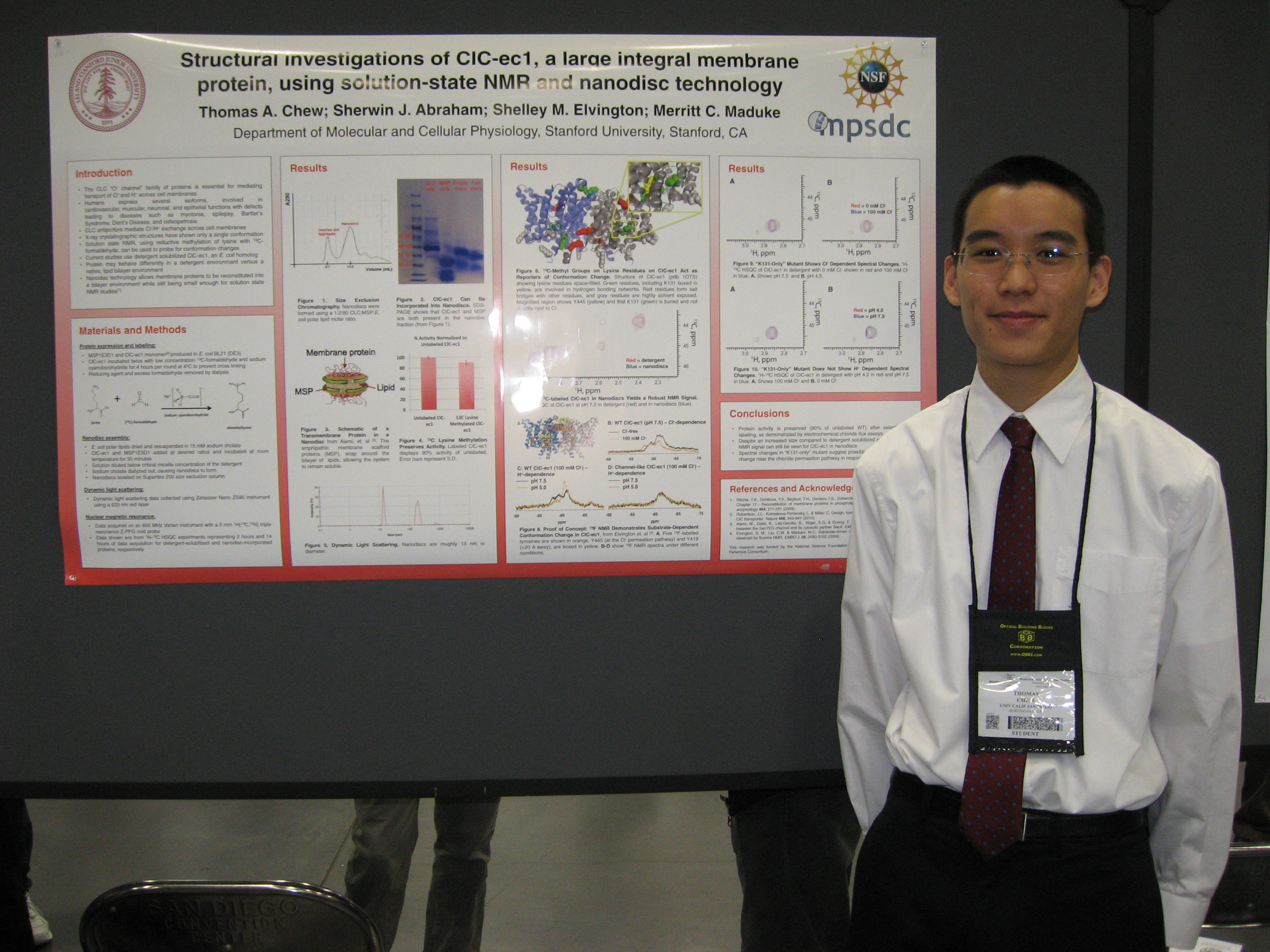
The winning poster is entitled Structural Investigations of CLC-ec1, A Large Integral Membrane Protein, Using Solution-State NMR and Nanodisc Techology and features Thomas Chew, Sherwin J. Abraham, Shelley M. Elvington, and Merritt Maduke as authors. The poster was selected by the National Institute of General Medical Sciences as the sole NIGMS Poster Winner.
 Thomas Chew at the February 27 Awards Ceremony together with the other winners and Steve Block. Click to enlarge.
Thomas Chew at the February 27 Awards Ceremony together with the other winners and Steve Block. Click to enlarge.Thomas was recognized and received a monetary award at the Awards Ceremony on February 27, preceding the 2012 National Lecture delivered by Steve Block. He was also selected to attend the NIGMS/NIH’s 50th anniversary symposium in October 2012.
Congratulations, Thomas and Merritt!
Read the Biophysical Society’s press release »


 Dr. Emad Tajkhorshid
Dr. Emad Tajkhorshid





