By Raymond E. Hulse
Department of Biochemistry and Molecular Biology, The University of Chicago
It is an understatement to say that the structural solution of the highly sought after voltage gated sodium channel is worth its weight in salt. Derived from a voltage gated bacterial sodium channel the solution, from Arcobacter butzleri (NaVAb), required a creative and demanding approach. The authors used techniques to achieve this structure that differ from other bacterial ion channels such as insect cell expression, the use of digitonin to solubilize and stabilize the protein, a lipid/detergent system for crystallization and strategic cysteine mutagenesis among several techniques. This achievement has been highly sought after since the discovery of bacterial voltage gated sodium channels and many researchers have spent considerable effort and time towards its elucidation.
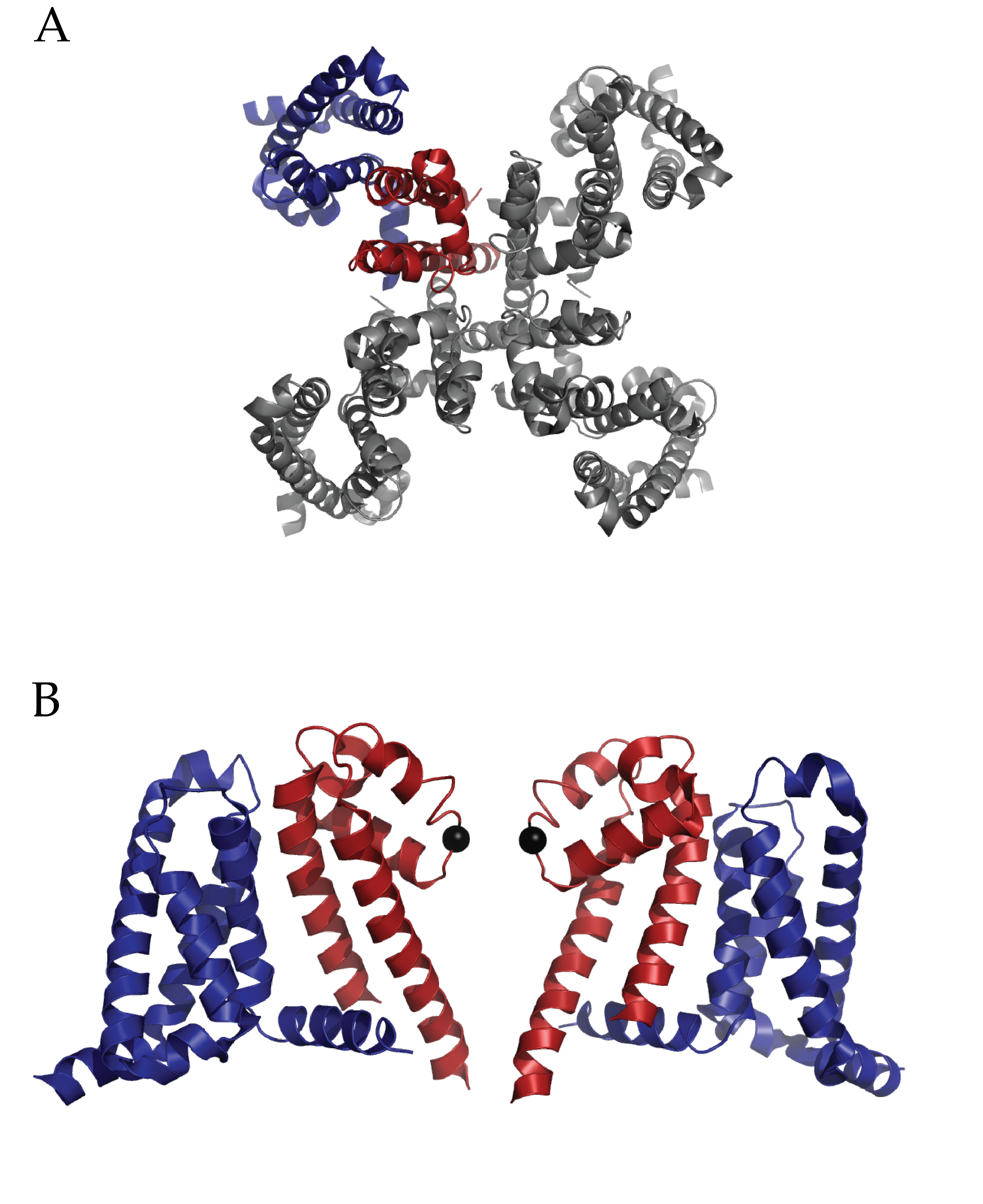
Figure 1 (click to enlarge)
Structure of Full length NaVAb: A) Extracellular top view of NaVAb. In dark blue is the VSD and in dark red is the PD of one monomer. B) A side view of two of four monomers (removed to ease visualization) of NaVAb. The alpha carbon of Glutamate 177 is represented as a black sphere. (PDB ID 3RVY)
Payandeh et al’s work will invigorate research into such critical questions as how Na+ ions are coordinated at the selectivity filter, alternative conformational states of a voltage sensor and the mechanism of voltage-dependent drug block via local anesthetics1. The pore domain of NaVAb is unique in that is possesses a helix-loop-helix conformation which deviates from well-researched potassium channels. The voltage sensor domain (VSD) also presents a surprise; the entire charge-sensing segment, S4, has an atypical secondary conformation of a 3-10 helix. Finally, side fenestrations in the central cavity of the pore domain are also present and provide a unique opportunity for further hypothesis driven research into pharmacological research. Notably the authors interpret their findings of this structure as a pre-activated voltage sensor with a closed pore.
One critical question surrounding this structure is the nature of coordination of Na+ ions. While potassium ion channels utilize the ability of the selectivity filter to dehydrate K+ ions, the Na+ ion is either incompletely dehydrated or not at all. The selectivity filter of voltage gated bacterial sodium channels is highly conserved. This structure demonstrates the role of those conserved sequences by revealing an intricate scaffold of hydrogen bond interactions between the selectivity filter and the two adjacent pore helices. Interestingly, these interactions occur within and between monomers. The structure does implicate the importance of one particular residue in the selectivity filter, a glutamate. The carboxylate group shares hydrogen bonds with nearby backbone amides on pore helices and the entire side chain forms the narrowest part of the pore. Yet there were no observed Na+ ions in the pore domain of the structure, which the authors ascribe to a low affinity.
In what is already a lively field of study, the conformational state of the VSD of NaVAb is striking. Foremost, the presence of a 3-10 helix along the sensing charges (R1-R4) in S4 is unique among all VSDs observed to date. Typically, a complete to partial alpha helix has been observed. An extensive network of interactions through the entire VSD is present. These include well-researched electrostatic and hydrophobic interactions with the charge sensor S4 in addition several structure specific types among the three other alpha helices (S1, S2 and S3). The authors interpret the structural information of the voltage sensing domain, in conjunction with previous disulfide locking experiments and comparison to other structures as evidence for an activated sensor. This is a remarkable finding considering the protein is not in the presence of an electric field and so not stimulated. Is it possible that the structure has trapped a different conformation such as an inactivated state? Functional assays on the particular cysteine mutations used to crystallize this structure may help further understanding.
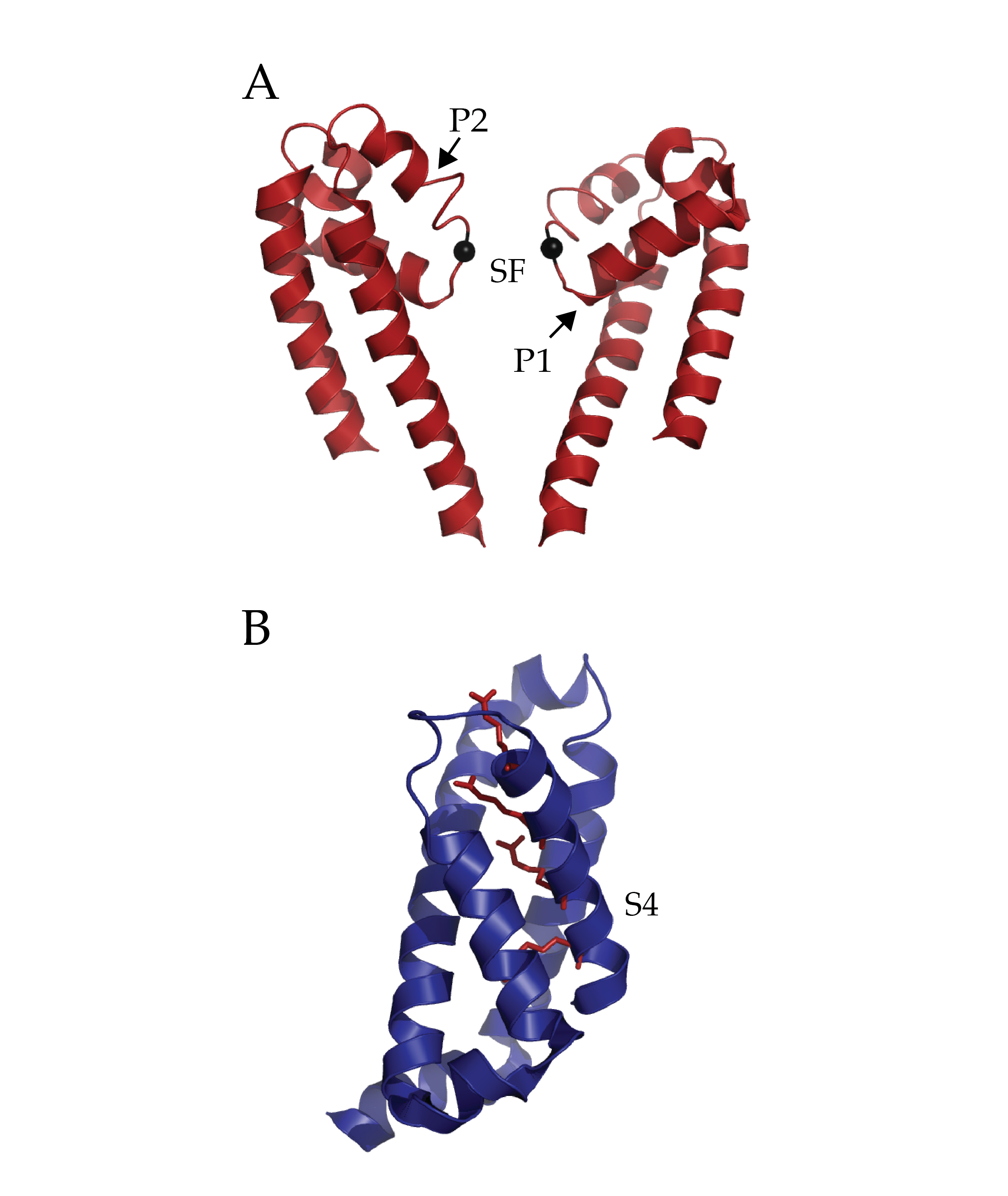
Figure 1 (click to enlarge)
Isolated Pore and Voltage Sensing Domain of NaVAb. A) Two of four pore domains isolated from remaining structure. The two pore helices are indicated with arrows (P1 and P2) and the selectivity filter (SF). The alpha-carbon of glutamate 177 in the selectivity filter has been rendered as a black sphere to ease localization. B) A single voltage sensing domain of NaVAb is depicted in cartoon format with the four sensing charges (R1-R4) of the S4 helix depicted in stick format (dark red). (PDB ID 3RVY)
Voltage gated sodium channels have long been researched in the context of pharmacology. This includes understanding the mechanism of pharmacological agents such as antiarrhythmics and local anesthetics (e.g., lidocaine). The side fenestrations of NaVAb are notable for their location; within the central cavity of the pore-domain itself. The presence of these windows gives the author an opportunity to present an interesting hypothesis: Are these the sites of use-dependent anesthesia block of sodium channels? The local hydrophobic environment might present a route for such drugs (e.g., etidocaine) to partition to the core of the membrane and then enter into the fenestration. The presence of a conserved phenylalanine residue at the fenestration with nearby conserved residues implicated in binding in eukaryotic channels is tantalizing. Ironically, while this family of bacterial voltage gated sodium channels select for Na+, they can be blocked by dihydropyridines (which block L-type Ca2+ channels). Future research into these mechanisms of pharmacological action will be enhanced greatly by this and future structures.
There are many interesting questions that arise from this work. How well does this particular channel represent both prokaryotic and eukaryotic voltage gated sodium channels? Bacterial voltage gated sodium channels are homotetrameric and so possess symmetry. Eukaryotic voltage gated sodium channels have a substantially more complex topology of a single, asymmetrical protein with 24 trans membrane helices. Additionally, kinetics of activation and inactivation differ between the two classes of channels. Sequence comparison of the growing family of bacterial voltage gated sodium channels to this new channel reveal several interesting differences. NaVAb lacks two glycines which have been noted for their role in gating for other bacterial voltage gated sodium channels2-4. NaVAb also appears to inactivate rapidly relative to other bacterial voltage gated sodium channels. Could this characteristic arise from minor sequence variations in the pore domain suggesting a potentially different mechanism of c-type inactivation or gating in this channel as has been observed in NaChBAC5? Finally how would mutations performed in earlier voltage-gated sodium channels in the selectivity filter promote Ca2+ selectivity6? Would such mutations alter the scaffold in the pore domain and change its stability and mechanism for selection?
Truly it is an exciting time for the sodium channel community in specific and the ion channel community in general. This work has great potential to help these communities understand the mechanism of how sodium channels work but also serve as a stepping stone towards understanding the nature of Ca2+ selectivity, perhaps the only topic more interesting than Na+ or K+ selectivity.
The structure of NaVAb has been rendered in cylinder format to facilitate visualization of the entire structure and rotated along the selectivity filter’s axis of symmetry. (PDB ID 3RVY)
Citations
1. Payandeh, J., Scheuer, T., Zheng, N. & Catterall, W.A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011).
2. Irie, K. et al. Comparative study of the gating motif and C-type inactivation in prokaryotic voltage-gated sodium channels. J Biol Chem 285, 3685–3694 (2010).
3. Zhao, Y., Scheuer, T. & Catterall, W.A. Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc Natl Acad Sci USA 101, 17873–17878 (2004).
4. O’reilly, A.O. et al. G219S mutagenesis as a means of stabilizing conformational flexibility in the bacterial sodium channel NaChBac. Mol Membr Biol 25, 670–676 (2008).
5. Pavlov, E. et al. The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys J 89, 232–242 (2005).
6. Yue, L., Navarro, B., Ren, D., Ramos, A. & Clapham, D. The Cation Selectivity Filter of the Bacterial Channel, NaChBac. Journal of General Physiology 1–9 (2002).
[/EXPAND]