Wednesday May 8th 2013, 9:00 AM to 5:00 PM
Gordon Center for Integrative Science, room W301/303
University of Chicago (directions)
Note: the Computational Modeling Core is also hosting a workshop specifically dedicated to membrane protein modeling at the University of Chicago on May 7th. Click here to learn more and register »
Force field and atomic models
| 9:00 | Lei Huang: Advances in automatic force field optimization |
| 9:15 | Christopher Mayne: Force Field Toolkit (FFTK) |
| 9:30 | Sunhwan Jo: Recent developments in CHARMM-GUI and CHARMM-GUI Membrane Builder |
| 9:45 | Wei Han: Further development of a mixed all-atom/united-atom models for simulation of peptide folding, aggregation and proteins in membrane |
| 10:00 | George Khelashvili: Protein-membrane coupling and the calculation of the bending rigidity of multi-component lipid membranes |
| 10:15 | Brigitte Ziervogel: Simulation of membrane systems with asymmetrical conditions |
| 10:30 | Josh Vermaas: Developing novel in silico solvents with optimized properties mimicking membrane models |
| 10:45 | Break |
| 11:00 | Niklaus Johner: Membrane proteins in the cubic phase lipid environment – insights into in meso crystallization environments |
| 11:15 | Yifei Qi: Quantifying the Drive-Response Relationships between Residues in Protein Folding |
Structural modeling with low-resolution data
| 11:30 | Aashish Adhikari: LBT loop modeling protocols |
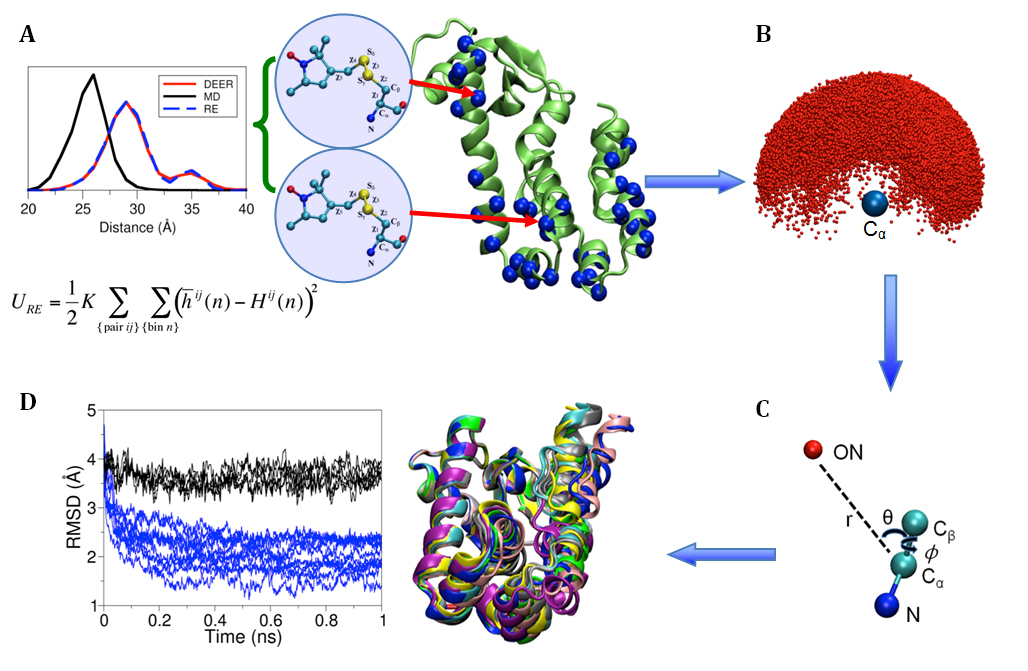
| 11:45 | Shahid Islam: Structural refinement using ESR/DEER histogram data |
| 12:00 | Fatemeh Khalili: Structural refinement using low-resolution LRET data |
| 12:15 | Lunch |
| 1:30 | Huan Rui: Probing Human Growth Hormone Receptor Structure Using Computational Simulations and NMR spectroscopy |
| 1:45 | Abhishek Singharoy: xMDFF – Molecular Dynamics Flexible Fitting of Low Resolution X-ray Structures |
| 2:00 | Hang Yu: Membrane Sculpting by F-BAR Domains by Molecular Dynamics Simulation |
Transition pathways
| 2:15 | Avisek Das: A simple method to determine conformational transition pathways based on a two-state anisotropic elastic network model |
| 2:30 | Mert Gur: Global Motions of Proteins Observed in Micro- to Milliseconds Simulations Concur with Anisotropic Network Model Predictions |
| 2:45 | Wen Ma: Study of conformational transitions of hexameric motors using the string method |
| 3:00 | Break |
| 3:15 | Mahmoud Moradi: Application of collective variables to describe large-scale conformational changes of membrane transporters |
| 3:30 | Zhe Wu: A New Hypothesis for the Mechanism of Synaptotagmin I in Synaptic Neurotransmitter Release |
| 3:45 | Po-Chao Wen: The impact of the lipid environment on gating of KcsA |
Spectroscopy
| 4:00 | Josh Carr: Introduction to computational analysis of 2D-IR spectroscopy |
Conclusion
| 4:15 | Roundtable discussion |