With our 2013 annual meeting less than a month away, we are delighted to share with you the news about a new computational simulation technique developed by several MPSDC team members that was first presented at last year’s Frontiers in Membrane Protein Dynamics conference. The development of this technique speaks to the significant scientific collaborations that take place under the umbrella of the Consortium, as well as the scientific conversations that began in Chicago last year.
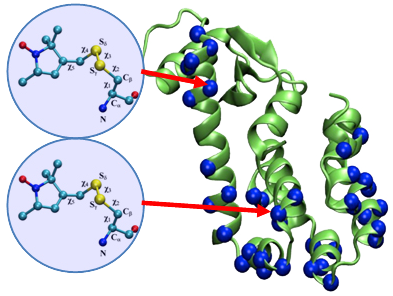
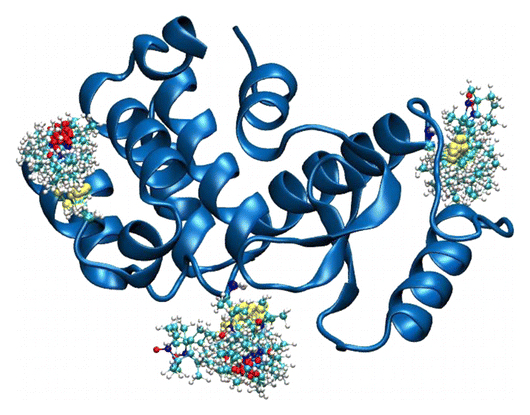 At the conference, Benoît Roux from our Computational Modeling Core introduced his team’s findings obtained from DEER (Double Electron-Electronic Resonance) data. At the conference, Roux and his team received helpful feedback from a number of scholars affiliated with the MPSDC as well as external invitees. After the conference, Roux and his team collaborated with a number of other scientists, including consortium colleague Hassane Mchaourab, to develop a novel computational simulation technique for exploiting the information from distance distribution data obtained from ESR/DEER spectroscopy for the refinement of membrane protein structures. This simulation technique, called the Restrained-Ensemble Molecular Dynamics (REMD) simulation method, uses a global ensemble-based energy restraint to force the spin-spin distance distribution histograms calculated from a multiple-copy molecular dynamics simulation to match those obtained from ESR/DEER experiments.
At the conference, Benoît Roux from our Computational Modeling Core introduced his team’s findings obtained from DEER (Double Electron-Electronic Resonance) data. At the conference, Roux and his team received helpful feedback from a number of scholars affiliated with the MPSDC as well as external invitees. After the conference, Roux and his team collaborated with a number of other scientists, including consortium colleague Hassane Mchaourab, to develop a novel computational simulation technique for exploiting the information from distance distribution data obtained from ESR/DEER spectroscopy for the refinement of membrane protein structures. This simulation technique, called the Restrained-Ensemble Molecular Dynamics (REMD) simulation method, uses a global ensemble-based energy restraint to force the spin-spin distance distribution histograms calculated from a multiple-copy molecular dynamics simulation to match those obtained from ESR/DEER experiments.
Already, the method has yielded three unique publications detailing the results of these experiments:
- Islam, S. M.; Stein, R.; Mchaourab, H.; Roux, B. Structural Refinement from Restrained-Ensemble Simulations Based on EPR/DEER Data: Application to T4 Lysozyme, J. Phys. Chem. B 117(17): 4740-54, 2013. (link)
- Roux, B.; Islam, S. M. Restrained-Ensemble Molecular Dynamics Simulations Based on Histograms from Double Electron-Electron Resonance Spectroscopy, J. Phys. Chem. B 117(17): 4733-9, 2013, In Press. (link)
- Roux, B.; Weare, J. On the statistical equivalence of restrained-ensemble simulations with the maximum entropy method, J. Chem. Phys. 138(8): 084107, 2013. (link)
 The article co-authored by Benoît Roux and Jeane Weare was highlighted by the Journal of Chemical Physics on their Top 20 Most Read in March 2013.
The article co-authored by Benoît Roux and Jeane Weare was highlighted by the Journal of Chemical Physics on their Top 20 Most Read in March 2013.
Roux and his team have also gone on to apply this method to VSD (voltage-sensing domain) data with Eduardo Perozo, and Glt(Ph) data with Olga Boudker. Additionally, Wonpil Im is also implementing an easy setup of this method with dummy spin-labels on his CHARMM-GUI generator.
Shahidul M. Islam from Roux’s team, who co-authored two of the above papers and has been deeply involved in the scientific process, provided the MPSDC with an overview of the technique and its utility. We invite you to read his overview here »
Congratulations to all involved in the development of this exciting and important new method!







