By Robert K. Nakamoto
Department of Molecular Physiology and Biological Physics, University of Virginia
There is almost nothing more useful to the biophysicist (except perhaps a talented grad student or post-doc) than to have probes in known positions within the structure of a protein. Spectroscopic and biochemical approaches use such probes to monitor the dynamics, environment and conformational shifts that occur during function. Probe placement is most commonly done by site-directed mutagenesis to introduce a unique cysteine and chemically modifying the purified protein with a sulfhydryl-reactive label. In recent years, investigators have developed new methods to incorporate custom-made amino acids directly into polypeptide chains by expanding the genetic code. In principle, amino acids with side chains of any chemical design can be incorporated into the growing polypeptide chain by the ribosome if the acyl-tRNA charged with the unnatural amino acid can be made. Usually a tRNA is used that recognizes one of the non-sense codons (UAG, AAG or UGA) or a codon not otherwise used in the coding sequence of the open reading frame. Of course, there are many limitations to this process but the main one is acylation of the 3’ base of the tRNA with the amino acid. This reaction is done in the cell by the amino acid-specific aminoacyl-tRNA synthetase. To broaden the genetic code, investigators have developed methods to “evolve” orthogonal synthetases to shift the specificities to their custom amino acid of choice (see Liu & Shultz for a review). Here, I describe a new and highly flexible method for synthesizing the acyl-tRNA with a wide range of unnatural amino acids using the “Flexizyme”.
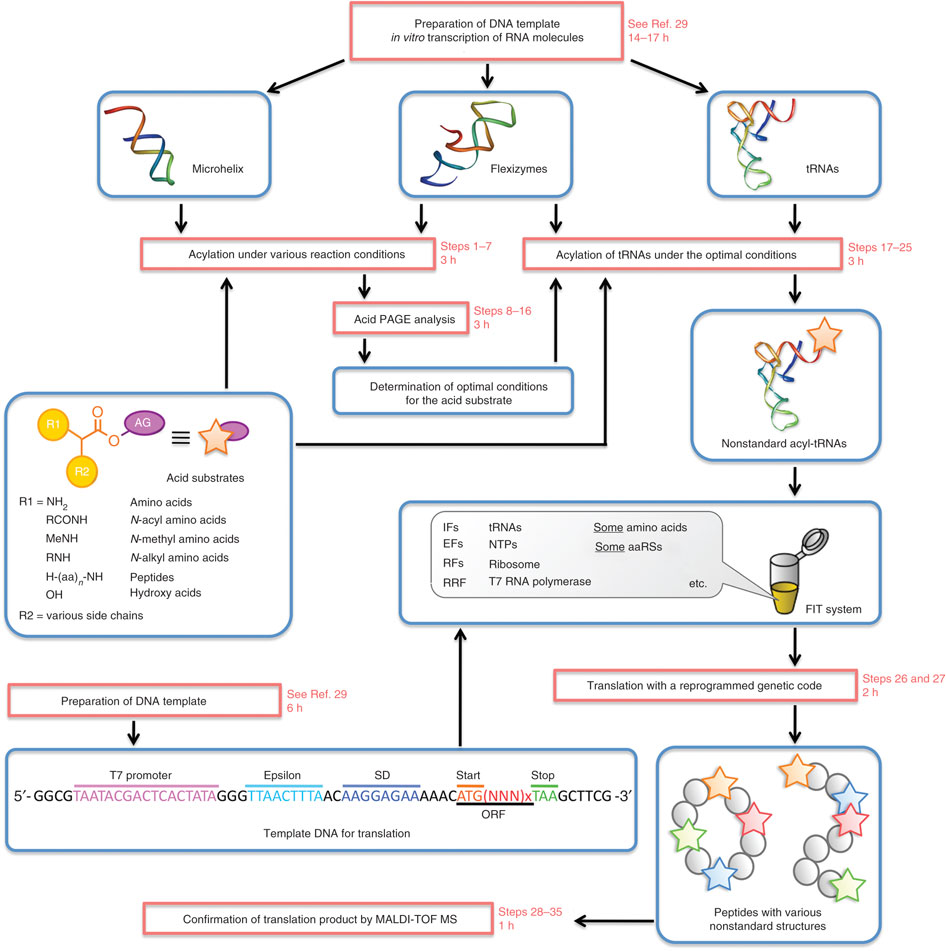
Developed and optimized in the laboratory of Professor Hiroaki Suga at the University of Tokyo, the “Flexizyme” approach is based on the use of a ribozyme to incorporate unnatural amino acids in proteins. A Nature Protocols paper (Goto et al.) describes the entire system and provides a detailed list of reagents and instructions. Though highly flexible in many ways, it is important to highlight that the system only works in cell-free translation systems.
Like in most systems designed to incorporate unnatural amino acids (see Liu & Shultz) a charged suppressor tRNA is used to integrate a designed amino acid into the nascent polypeptide chain. However, rather than acylating the tRNAsup with an aminoacyl-tRNA synthetase specifically evolved to accept only a given unnatural amino acid, the Flexizyme system acylates the tRNAsup with a ribozyme. This avoids altogether the difficult and somewhat daunting process of evolving an orthogonal tRNA synthetase to recognize each unnatural amino acid. The Suga laboratory has developed a set of Flexizymes capable of charging a tRNA with a diverse array of unnatural amino acids. Because the system is used only in vitro, it also avoids the additional worry of importing the unnatural amino acids into a cell, which can have toxic effects.
A key part of the system is the acyl-donor substrate, which is used by the Flexizyme to acylate the tRNAsup. Because the reaction is primarily driven by the nature of the leaving group, the amino acid part of the substrate can vary. Suga and his colleagues have developed a set of three Flexizymes optimized to work with four different leaving groups. The Flexizyme system is particularly attractive for incorporating spectroscopic probes such as fluorescence or nitroxide spin labels at user-defined positions. Normally, investigators try to create a background devoid of cysteines and unique sulfhydryls are introduced to provide specific reactive positions. The Flexizyme system abrogates the potential problems that replacing native Cys can have on protein stability and function, or that a given Cys may be inaccessible and unreactive.The investigator must determine which donor-leaving group/Flexizyme combination works best with their specific unnatural amino acid. For example, aromatic side chains generally work best with the cyanomethyl ester-leaving group and the “eFx” ribozyme, while non-aromatic side chains would use the 3,5-dinitrobenzyl ester with the “dFx” ribozyme (see Goto et al. for sequences and structures). The Flexizymes and tRNAs are easily made in the laboratory and the chemistries to synthesize the acyl-donor substrates are relatively straightforward. Assays to determine the efficiencies of the transfer reactions are described by Murakami et al.
In a very nice example, Öjemalm et al. used the Flexizyme system to create substrates to study the influence of polypeptide hydrophobicity on insertion into the endoplasmic reticulum membrane via the Sec61 translocon. The Flexizyme was used to charge the tRNAsup with a series of unnatural amino acids with linear aliphatic side chains up to 10 carbons in length.
The Flexizyme system is particularly attractive for incorporating spectroscopic probes such as fluorescence or nitroxide spin labels at user-defined positions. Normally, investigators try to create a background devoid of cysteines and unique sulfhydryls are introduced to provide specific reactive positions. The Flexizyme system abrogates the potential problems that replacing native Cys can have on protein stability and function, or that a given Cys may be inaccessible and unreactive. Furthermore, there is a wider choice of labels with the Flexizyme and the investigator can use the proper probe to monitor specific properties such as backbone dynamics or local environments, or to impart desired modifications into the structure.
In combination with robust cell free synthesis systems such as those developed by Volker Dõtsch and colleagues (see Klammt et al. for a review, and the Membrane Protein Expression and Purification Core page of this website for detailed protocols), the Flexizyme system can be a powerful tool for the efficient production of the modified protein in biochemical amounts. Obviously, the system is potentially limited by the ease with which the in vitro synthesized polypeptide can be folded into its native conformation. This can be particularly difficult with integral membrane proteins, since folding must be done in detergents, lipids or a complex combination of both. Recent successes are indeed encouraging and the advantages provided by combining the two approaches represent an untapped resource in the analysis of the structure and dynamics of membrane proteins.
References
Goto, Y., Katoh, T. and Suga, H. (2011) Flexizymes for genetic code reprogramming. Nature Protoc. 6, 779-790.
Klammt, C., Löhr, F., Schäfer, B., Haase, W., Dötsch, V., Rüterjans, H., Glaubitz, C. and Bernhard, F. (2004) High level cell-free expression and specific labeling of integral membrane proteins. Eur. J. Biochem. 271, 568–580.
Liu, C. C. and Schultz, P. G. (2010) Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413-444.
Murakami, H., Ohta, A., Ashigai, H. and Suga, H. (2006) A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nature Meth. 3, 357-359.
Öjemalm, K., Higuchi, T., Jiang, Y., Langel, Ü., Nilsson, I., White, S. H., Suga, H. and von Heijne, G. (2011) Apolar surface area determines the efficiency of translocon-mediated membrane-protein integration into the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 108, 359-364.
[/EXPAND]