Dr. Klaus Schulten
Dr. Klaus Schulten
Three weeks in advance of Frontiers in Membrane Protein Structural Dynamics 2014, Professor Klaus Schulten (Director of the Computational and Theoretical Biophysics Group at UIUC) phoned in with us from his native Germany to talk about his research interests, the state of the field, the Membrane Protein Structural Dynamics Consortium (MPSDC), and the keynote lecture that he will be giving at the conference.
![]() The full movie that will be shown in Klaus Schulten’s lecture can be viewed here.
The full movie that will be shown in Klaus Schulten’s lecture can be viewed here.
Interviewer:
This is Rudo Kemper conducting an interview with Professor Klaus Schulten from the University of Illinois at Urbana-Champaign for the Membrane Protein Structural Dynamics Consortium.
Professor Schulten, please describe some of your scientific research interests, both past and present.
Klaus:
I’m a theoretical biophysicist and I try to approach living cells as a physicist to explore in how far cells exploit physical properties and physical laws and thereby to understand them better. My long goal is to achieve this by using theory as a kind of microscope and zoom through computer simulation into the processes in living cells; the views achieved this way are unobtainable for experiment, but in many ways can be verified by experiment. In other words, I wanted to build the world’s best microscope to view living cells and I sort of achieved it. Like any microscope, you can improve the computational microscope and reduce imaging artifacts, but by and large we can today image living cells through computer simulation with very high resolving power.
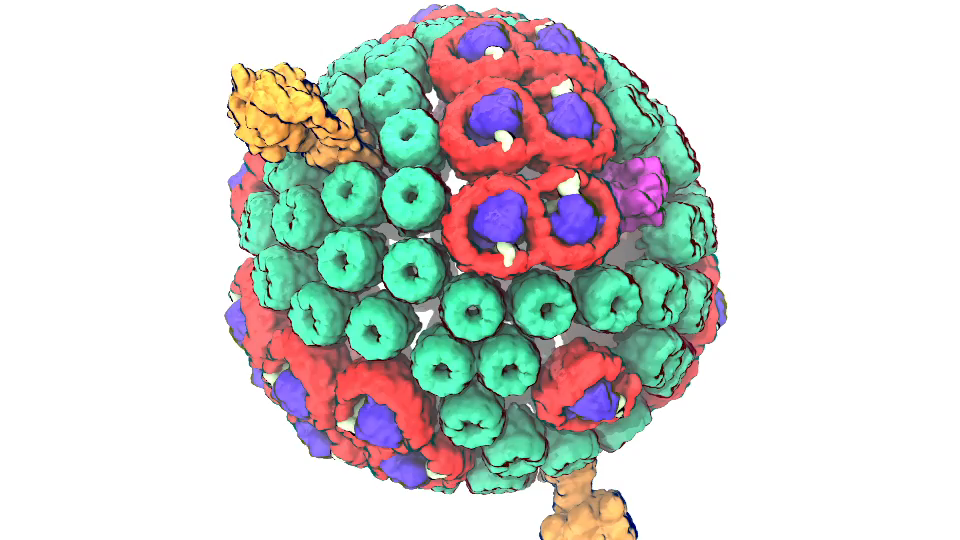 Photosynthetic chromatophore organelle (click for movie)
Photosynthetic chromatophore organelle (click for movie)
Interviewer:
In your view, what are some of the key challenges for the field of membrane protein biophysics in the coming 5-10 years?
Klaus:
One shouldn’t limit the question just to membrane biophysics because the membrane is only one part of the living cell. The cell has many components, the membrane is just one, and one has to see the cell as a whole rather than just focusing on one part. looking at small parts of the cell has been the dominant research focus for the last 50 years, looking basically at one protein at a time. In fact, biophysics had been extremely focused inside the cell looking at a volume that measured about 100 Angstroms x 100 Angstroms x 100 Angstroms at best, containing single proteins or protein complexes. But this cell volume is not where a biological cell really becomes alive.
The smallest living entity in Nature is a cell. A cell is 1 micrometer to 10 micrometers long in every direction, and so it is much bigger than the volume taken by single proteins or protein complexes. But somewhere between the small volume of a single protein and the large volume of the whole cell, life comes about. And this happens is what we want to understand, and we want to see what is it that makes life possible, that a cell can reproduce, can repair itself, organize itself, defend itself, live optimally in its environment. The point is actually rather simple: when you increasing the size of the volume of the cell that you visualize and thereby investigate, you may see at some volume size a very important phenomenon, namely how the many single molecules begin to cooperate in the cell and bring about the state that we call living. To appreciate this one needs to note that a living cell, for example a human cell, contains as many proteins as the United States has citizens. And so as have in our country many forms of organization, starting with the individual, then the family, then the neighborhood, the village, the city, the state and the country; there are also shops, companies, factories an army and more. And so likewise "social" interactions are also critical for the living cell and, in fact, it is the association and cooperation of molecules, for example proteins, what makes a cell alive. And so if you want to study how the living state develops, you have to study a volume of the cell that is much bigger than just what was studied before. Before, we studied phenomena in 100 Angstroms x 100 Angstroms x 100 Angstroms volume that contain up to about one million atoms. In the new era we seek to study volumes that contain up to a billion atoms. To visualize this large volume computationally we need to describe the detailed motion of as many atoms. That is a great challenge, but worth pursuing as we can learn then how biomolecules associate and cooperate in the cell, organize themselves, and thereby bring about life as we know it.
Interviewer:
What are you planning to talk about in your keynote lecture during the Frontiers in Membrane Proteins Structural Dynamics Conference in May?
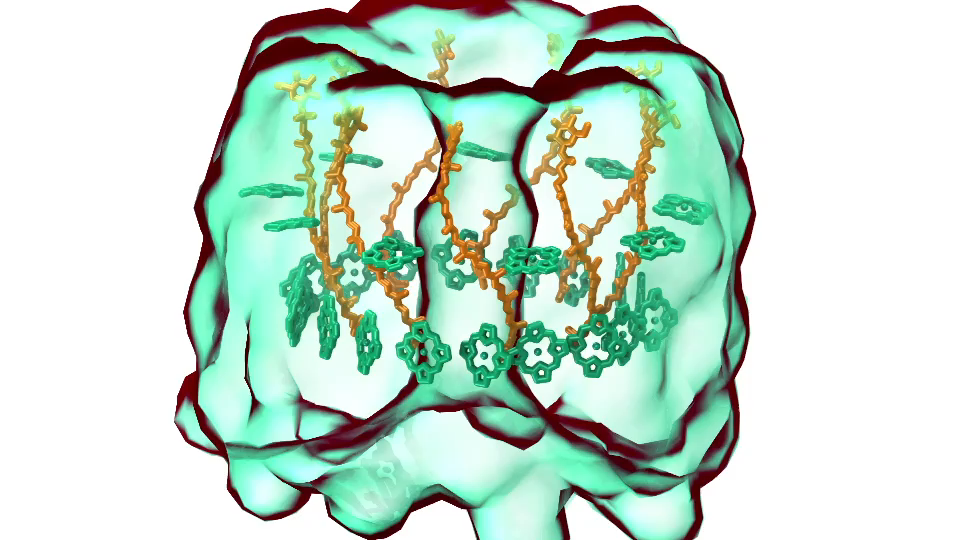 LH1 – reaction center complex (click for movie)[caption align="alignright" width="300"]
LH1 – reaction center complex (click for movie)[caption align="alignright" width="300"]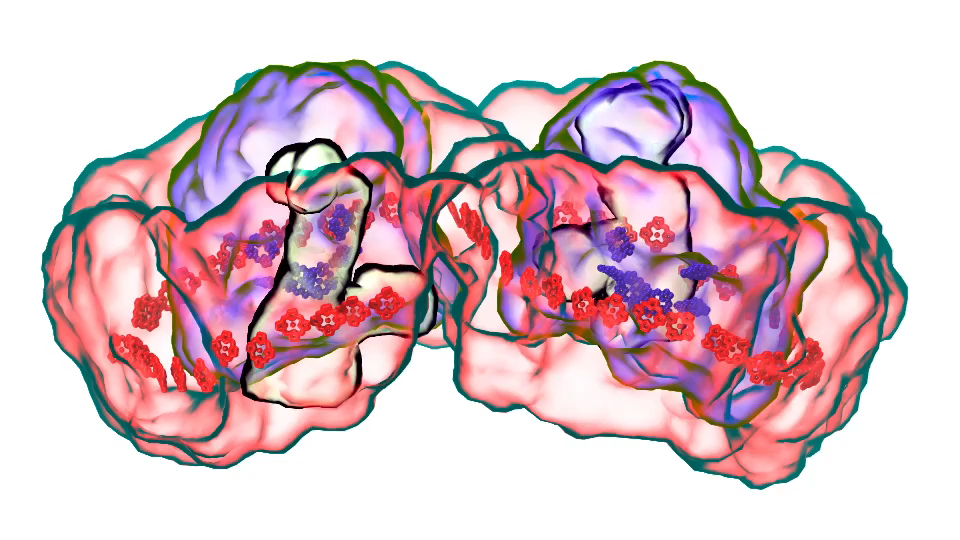 LH2 complex (click for movie)[/caption]
LH2 complex (click for movie)[/caption]
Klaus:
In Chicago I will talk about exactly one of those cases that I just mentioned. My talk is about a membrane system, called a chromatophore, that contains over a 100 million atoms a volume 700 Angstroms x 700 Angstroms x 700 Angstroms size. The chromatophore is a spherical membrane and will describe the processes that organize themselves in the chromatophore. The chromatophore is also called a cellular organelle and is found in huge numbers, like several hundreds, in certain photosynthetic bacteria, called purple bacteria. The chromatophore is a very critical organelle for the bacterium, because it gives the bacterium the energy it needs to sustain its life. It enables the bacterium to absorb sunlight and to turn it into chemical fuel that drives many processes in the cell. The key point of my lecture is that I will present the chromatophore as a whole. In the past we had been looking only at bits and pieces, at a single protein here, a light absorption process there. Now I show in my lecture that it is possible today to study the chromatophore, made of hundreds of proteins that permit many processes to run their course, as an entire system with all parts and processes included.
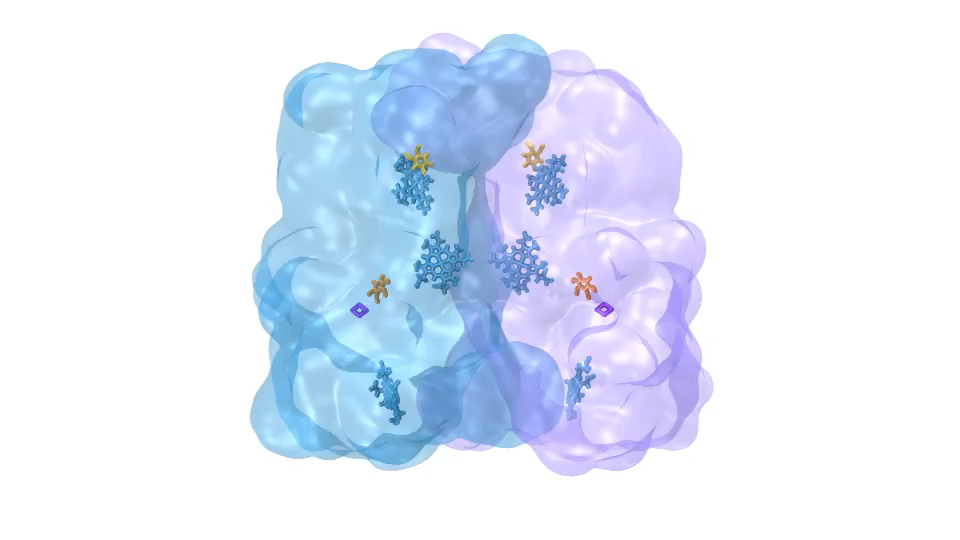 ATP synthase (click for movie)[caption align="alignright" width="300"]
ATP synthase (click for movie)[caption align="alignright" width="300"]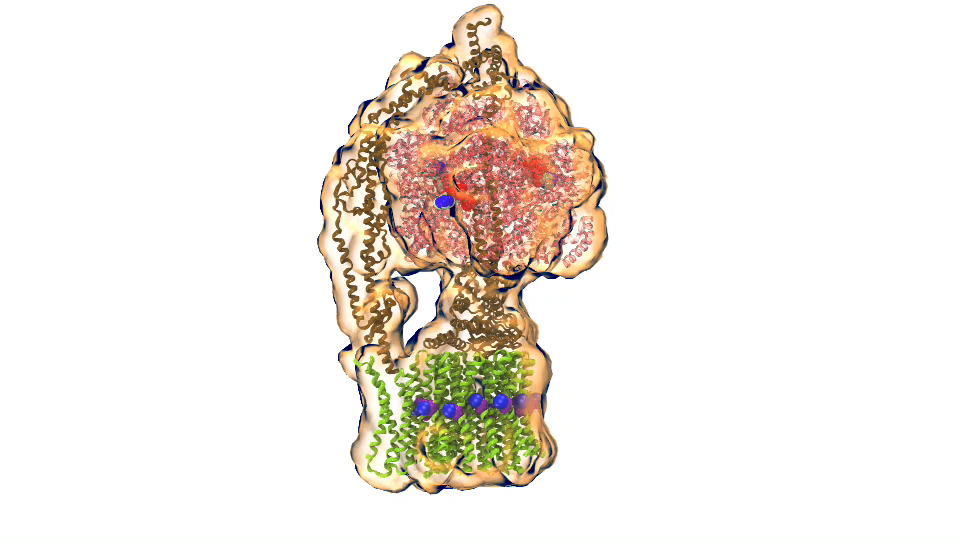 bc1 complex (click for movie)[/caption]
bc1 complex (click for movie)[/caption]
I had to actually work for 37 years of my life to get where I am and what I present in my lecture, and I’m not quite at the end even after so many years, but came far to give my lecture. The lecture will describe how all processes interlock with each other very much like the wheels making the horary in a Swiss watch. The processes (wheels) are all functioning together and work very precisely, just like a watch, despite thermal noise and other natural disorder. The very large biomolecular association that is the chromatophore has its architecture and processes perfectly attuned to permit purple bacteria that live mostly in very dim light, to nevertheless namely to make a living from the very few photons that exist in their habitat.
The chromatophore is composed of about 200 proteins and carries out about 30 processes. My lecture will start and end with an atom-by-atom movie, that is actually quite beautiful just like a movie in the theater, where one see all these processes taking place from the beginning when a photon is absorbed, all the way to the end when one sees the cellular fuel, molecules of ATP, being synthesized. One sees how the processes in the chromaotphore drive each other like a clockwork and are arranged beautifully together.
Interviewer:
In your view, how does the Glue Grant funded Membrane Protein Structural Dynamics Consortium contribute to the field?
Klaus:
The Consortium is contributing in two main respects to the field of membrane biology. On one side, it provides tools and laboratory capacities for membrane research. Membrane research is really very important, a key part of the cell is actually the membrane, many processes take place there, but it is a part of the cell that is much harder to study than the main part of the cell, called the cytosol. The Consortium develops tools that many other scientists can use, and it also provides collaborative capacities, so that researchers outside the Consortium can use the tools, coming to visit researchers who are members of the consortium, to collaborate with them and use their laboratory facilities.
This is one respect. The other respect is doing the most advanced research in membrane biology. For this purpose the consortium is constantly improving research tools so that the Consortium remains at the forefront of the field. Basically the mission of the Consortium is simple: do great science and at the same time develop new tools and make those tools available to everybody. This is a very, very important mission. The Consortium has a particularly strong computational side and, in keeping with its mission develops software that is used by many researchers in the United States and the world. The computational Consortium members have many researchers coming to visit for the investigation of membrane processes.
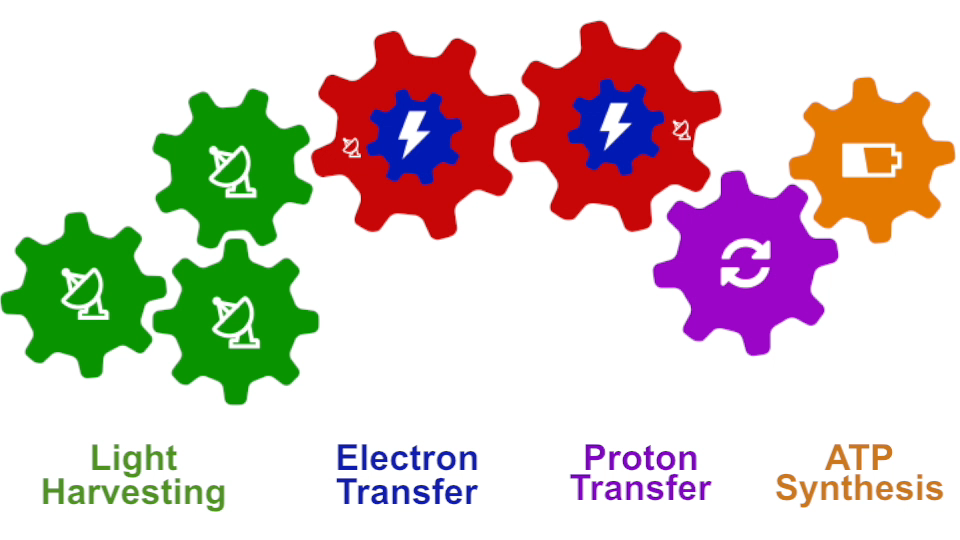 Photosynthesis – just like clockwork (click for movie)
Photosynthesis – just like clockwork (click for movie)
Interviewer:
Finally, how have you and your team collaborated with some of the ongoing MPSDC projects?
Klaus:
My laboratory style is that basically all science projects are done together with experimentalists. As a result I benefit tremendously from the excellent experimental colleagues of the consortium. In the Consortium I have also really outstanding computational colleagues with whom I can develop new tools.
To be specific, we have one particular project where we study what is called the voltage sensing domain1. It is a very critical part of the so-called ion channels that exist, for example, in nerve cells, that have to adapt themselves to the spontaneous voltage of the nerve cell membrane. In this particular case, the experimentalists initially didn’t have the resolution power to get from their experimental observation the full atomic picture of the voltage sensing domain. But with a tool that my group developed for the purpose and tried out for the first time in this collaboration, we could actually develop a computationally resolve a key part of the atomic level structure. Later the experimentalist could actually solve the structure themselves, and we had a perfect agreement with the computational results so that we published together. We could also, as modelers, begin to explore the function of the voltage sensing domain. In any case we had a very close collaboration between computational and experimental membrane biologists.
1. Schulten has recently collaborated with the laboratories of Consortium colleagues Eduardo Perozo, Benoit Roux, and Anthony Kossiakoff to publish a paper titled Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain – an F1000Prime recommended article published in Nature Structural & Molecular Biology in March 2014
Klaus Schulten is Professor of Biophysics and Director of the Computational and Theoretical Biophysics Group at University of Illinois at Urbana-Champaign. Schulten is a leader in the field of computational biophysics, having devoted over 40 years to establishing the physical mechanisms underlying the processes and organization of living systems, from the atomic scale up to the level of the entire organism. As of 2014, his work in biological physics has yielded over 625 publications, which have been cited over 67,000 times. Schulten is also the co-director of the NSF-funded Center for the Physics of Living Cells and his work has been honored with numerous awards, including the Distinguished Service Award of the Biophysical Society in 2013, and the IEEE Computer Society Sidney Fernbach Award in 2012. Schulten also received the prestigious Humboldt Award of the German Humboldt Foundation in 2004.
Interview by Rudo Kemper, Web Content Administrator, Information and Dissemination Core