Dr. Shohei Koide‘s collaborations in a scientific team seeking to discover the structure of a fluoride-specific ion channel was recently featured in Brandeis NOW, a scientific research publication published by Brandeis University. The work focuses on Dr. Christopher Miller’s lab, based at Brandeis University. The research was supported in part by the Consortium, and culminated in the following publication:
Researchers discover structure of fluoride-specific ion channel
Creation of ‘atomic blueprint’ is a biological novelty
The original Brandeis NOW press release by Kimm Fesenmaier can be read here
 Fluoride protects our teeth against cavity-causing bacteria by making our teeth stronger. But what if we could find a way to trap fluoride ions (the negatively charged form of the chemical element fluorine) inside bacteria? At the right concentration, fluoride ions are highly toxic to bacteria, wreaking havoc on their proteins and disrupting critical cellular functions. Bacteria, however, can fight back, exporting the toxic fluoride ions out using specialized proteins called fluoride-specific ion channels.
Fluoride protects our teeth against cavity-causing bacteria by making our teeth stronger. But what if we could find a way to trap fluoride ions (the negatively charged form of the chemical element fluorine) inside bacteria? At the right concentration, fluoride ions are highly toxic to bacteria, wreaking havoc on their proteins and disrupting critical cellular functions. Bacteria, however, can fight back, exporting the toxic fluoride ions out using specialized proteins called fluoride-specific ion channels.
How these proteins remove fluoride ions from the cell is poorly understand. To glimpse into the inner workings of these proteins, researchers in Christopher Miller’s lab at Brandeis University in collaboration with Simon Newstead at the University of Oxford have determined the structure of one such fluoride-specific ion channel from the Bordetella pertussis bacteria called Bpe.
In a paper published by Nature on Sept. 7, lead author Randy Stockbridge, a post-doctorate fellow in Miller’s lab, and colleagues used a technique called X-ray crystallography to obtain an “atomic blueprint” illustrating the arrangement of the amino acids that make up Bpe. The details provide important insight into how Bpe exports fluoride out of the cell. Intriguingly, the schematics also point out potential weaknesses in Bpe that could be exploited to trap fluoride inside bacteria.
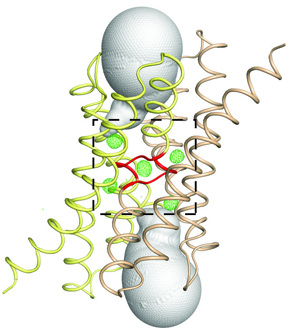
Based on the blueprint, the Bpe fluoride-specific ion channel resembles an hourglass and is actually composed of two Bpe protein molecules. At the center where the hourglass constricts is a sodium ion that may act like a pin that fastens the two Bpe proteins together. However, rather than having a central pore like an hourglass, the arrangement of the two Bpe molecules forms two parallel tunnels through which fluoride ions could flow. The blueprint also helps explain why Bpe only exports fluoride; the tunnels are just the right size for fluoride ions, but are too narrow for the biologically abundant chloride ion, fluoride’s larger but chemically similar cousin.
Notably, the researchers observed two unidentifiable “hazy shadows” in each tunnel, which they concluded were fluoride ions. First, many of the amino acids along the walls of each channel that protrude toward the shadows are chemically attracted to fluoride ions. Consistently, when the researchers mutated some of these amino acids to change their chemical properties, Bpe’s ability to export fluoride ions out of the bacteria was dramatically reduced. The researchers also studied the structure of a related fluoride-specific ion channel, Ec2, and found similar hazy shadows in its tunnels.
The researchers also noted a peculiar feature of Bpe that has implications for how the protein moves fluoride out. At one point in each of the tunnels, the side chain, or chemical appendage, of a particular amino acid protrudes inward, contacts the fluoride ion, and impedes its path to the exit. The side chain can swivel though, so it may be that the fluoride ion grasps it as though it were a turnstile and is then pulled to the other side when the side chain rotates.
Armed with Bpe’s blueprints, researchers can exploit structural weaknesses and develop strategies that kill bacteria by preventing fluoride from being moved out. Chemical compounds could be designed that pull the sodium ion pin and dismantle Bpe or that lock the turnstile causing a fluoride traffic jam that backs up into the cell.
In addition to Stockbridge, Miller and Newstead, the paper’s other authors are Ludmila Kolmakova-Partensky, Tania Shane, Akiko Koide and Shohei Koide.
The research was supported in part by a Wellcome Trust Investigator Award and grants from the National Institutes of Health (NIH) (RO1-GM107023 and U54-GM087519). Stockbridge also was supported by an NIH grant (K99-GM-111767). Miller is a Howard Hughes Medical Investigator.





 Not just anything is allowed to enter the nucleus, the heart of eukaryotic cells where, among other things, genetic information is stored. A double membrane, called the “nuclear envelope,” serves as a wall, protecting the contents of the nucleus. Any molecules trying to enter or exit the nucleus must do so via a cellular gatekeeper known as the nuclear pore complex (NPC), or pore, which exists within the envelope.
Not just anything is allowed to enter the nucleus, the heart of eukaryotic cells where, among other things, genetic information is stored. A double membrane, called the “nuclear envelope,” serves as a wall, protecting the contents of the nucleus. Any molecules trying to enter or exit the nucleus must do so via a cellular gatekeeper known as the nuclear pore complex (NPC), or pore, which exists within the envelope. Fluoride protects our teeth against cavity-causing bacteria by making our teeth stronger. But what if we could find a way to trap fluoride ions (the negatively charged form of the chemical element fluorine) inside bacteria? At the right concentration, fluoride ions are highly toxic to bacteria, wreaking havoc on their proteins and disrupting critical cellular functions. Bacteria, however, can fight back, exporting the toxic fluoride ions out using specialized proteins called fluoride-specific ion channels.
Fluoride protects our teeth against cavity-causing bacteria by making our teeth stronger. But what if we could find a way to trap fluoride ions (the negatively charged form of the chemical element fluorine) inside bacteria? At the right concentration, fluoride ions are highly toxic to bacteria, wreaking havoc on their proteins and disrupting critical cellular functions. Bacteria, however, can fight back, exporting the toxic fluoride ions out using specialized proteins called fluoride-specific ion channels.





